MCMC sampling
Marchov Chain Monte Carlo sampling- draw samples proportional to likelihood

- two parameters $\alpha$ and $\beta$
- infinitely vague priors - posterior likelihood only
- likelihood multivariate normal
> modelString="
+ model {
+ #Likelihood
+ for (i in 1:n) {
+ y[i]~dnorm(mu[i],tau)
+ mu[i] <- beta0+beta1*x[i]
+ }
+
+ #Priors
+ beta0 ~ dnorm (0,1.0E-6)
+ beta1 ~ dnorm(0,1.0E-6)
+ tau <- 1 / (sigma * sigma)
+ sigma~dunif(0,100)
+ }
+ "
> writeLines(modelString,con="BUGSscripts/model1.txt")
> data.list <- with(data, list(y = Y, x = X, + n = nrow(data))) > data.list
$y [1] 1.374 3.684 4.164 8.095 8.330 [6] 8.680 11.487 $x [1] 0 1 2 3 4 5 6 $n [1] 7
> params <- c("beta0", "beta1", "sigma") > adaptSteps = 1000 > burnInSteps = 2000 > nChains = 3 > numSavedSteps = 50000 > thinSteps = 1 > nIter = ceiling((numSavedSteps * thinSteps)/nChains)
> library(R2jags) > data.r2jags <- jags(data=data.list, + inits=NULL, + parameters.to.save=params, + model.file="BUGSscripts/model1.txt", + n.chains=3, + n.iter=nIter, + n.burnin=burnInSteps, + n.thin=thinSteps + )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 38 Initializing model
> plot(as.mcmc(data.r2jags))
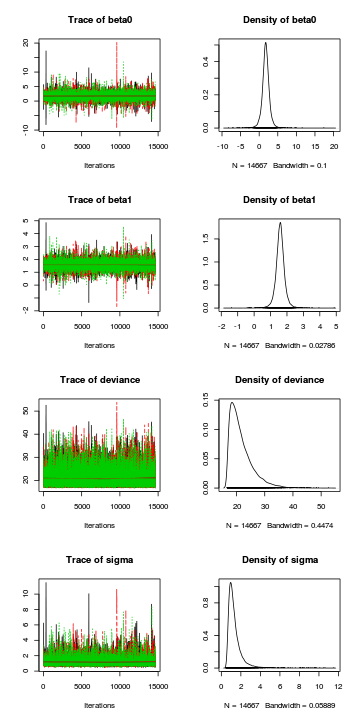
> raftery.diag(as.mcmc(data.r2jags))
[[1]]
Quantile (q) = 0.025
Accuracy (r) = +/- 0.005
Probability (s) = 0.95
Burn-in Total Lower bound
(M) (N) (Nmin)
beta0 3 4586 3746
beta1 4 7366 3746
deviance 3 4054 3746
sigma 4 4910 3746
Dependence
factor (I)
1.22
1.97
1.08
1.31
[[2]]
Quantile (q) = 0.025
Accuracy (r) = +/- 0.005
Probability (s) = 0.95
Burn-in Total Lower bound
(M) (N) (Nmin)
beta0 9 13827 3746
beta1 6 8180 3746
deviance 3 4008 3746
sigma 4 4773 3746
Dependence
factor (I)
3.69
2.18
1.07
1.27
[[3]]
Quantile (q) = 0.025
Accuracy (r) = +/- 0.005
Probability (s) = 0.95
Burn-in Total Lower bound
(M) (N) (Nmin)
beta0 6 8440 3746
beta1 6 8192 3746
deviance 2 3919 3746
sigma 4 5081 3746
Dependence
factor (I)
2.25
2.19
1.05
1.36
> autocorr.diag(as.mcmc(data.r2jags))
beta0 beta1 deviance
Lag 0 1.000000 1.000000 1.00000
Lag 1 -0.008001 -0.001116 0.66897
Lag 5 0.012437 0.002244 0.26645
Lag 10 0.003184 0.005771 0.11655
Lag 50 0.001979 0.004254 0.00254
sigma
Lag 0 1.000000
Lag 1 0.793155
Lag 5 0.390997
Lag 10 0.203675
Lag 50 0.001013
> print(data.r2jags)
Inference for Bugs model at "BUGSscripts/model1.txt", fit using jags,
3 chains, each with 16667 iterations (first 2000 discarded)
n.sims = 44001 iterations saved
mu.vect sd.vect 2.5% 25%
beta0 1.770 1.045 -0.302 1.238
beta1 1.590 0.287 1.023 1.440
sigma 1.348 0.721 0.633 0.915
deviance 21.650 4.111 17.057 18.667
50% 75% 97.5% Rhat
beta0 1.773 2.311 3.811 1.001
beta1 1.590 1.739 2.163 1.001
sigma 1.161 1.546 3.226 1.001
deviance 20.539 23.467 32.494 1.001
n.eff
beta0 38000
beta1 44000
sigma 44000
deviance 44000
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 8.5 and DIC = 30.1
DIC is an estimate of expected predictive error (lower deviance is better).