Poisson regression
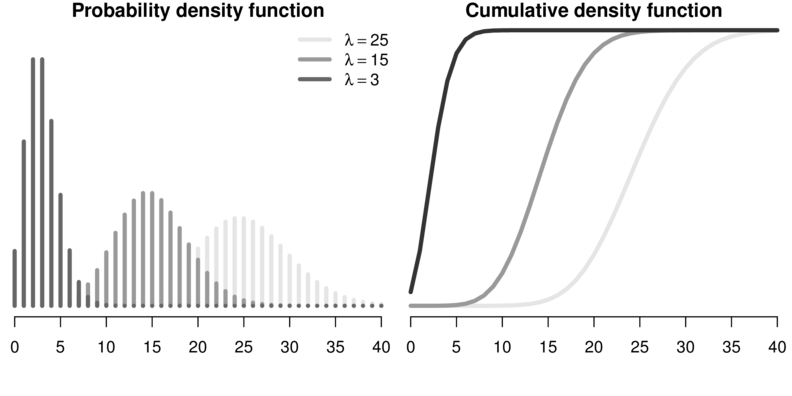
\[
p(Y_i) = \frac{e^{-\lambda}\lambda^x}{x!}
\]
\[log(\mu)=\beta_0+\beta_1x_i+...+\beta_px_p\]

Spread assumed to be equal to mean. (\(\phi = 1\))

Sample more varied than expected from its mean
Sample more varied than expected from its mean
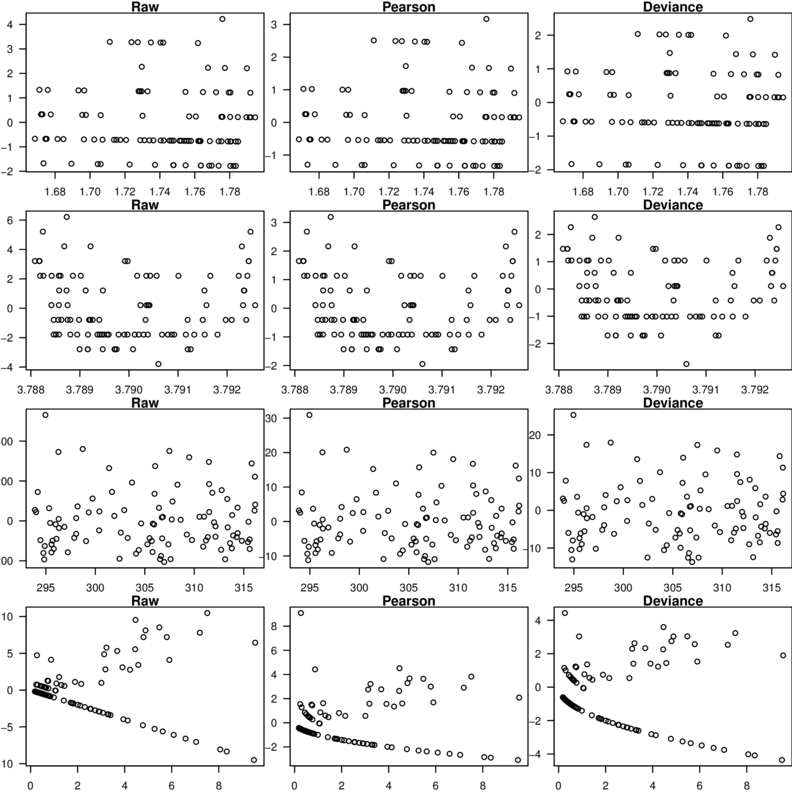
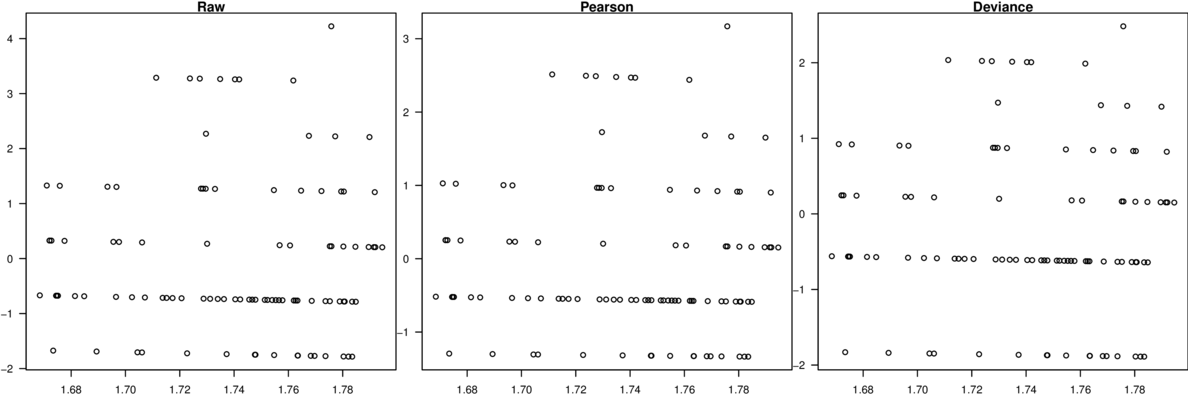

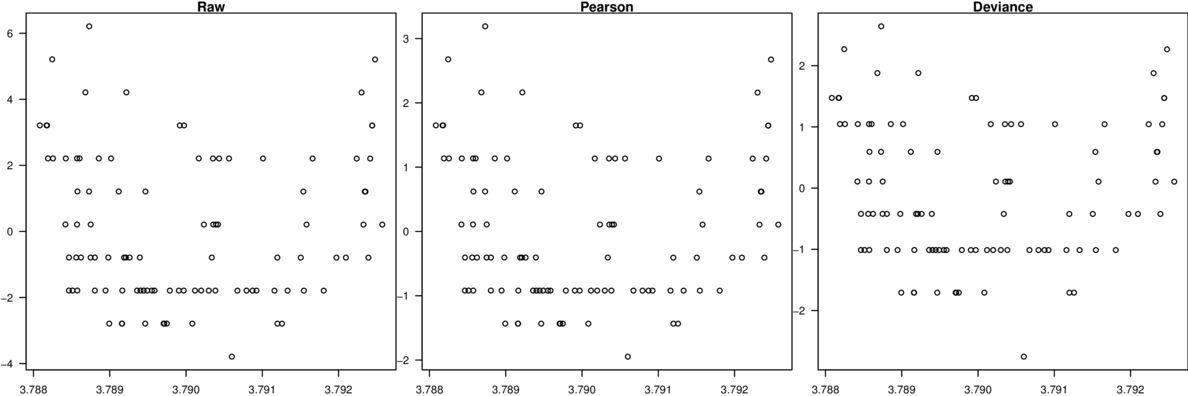
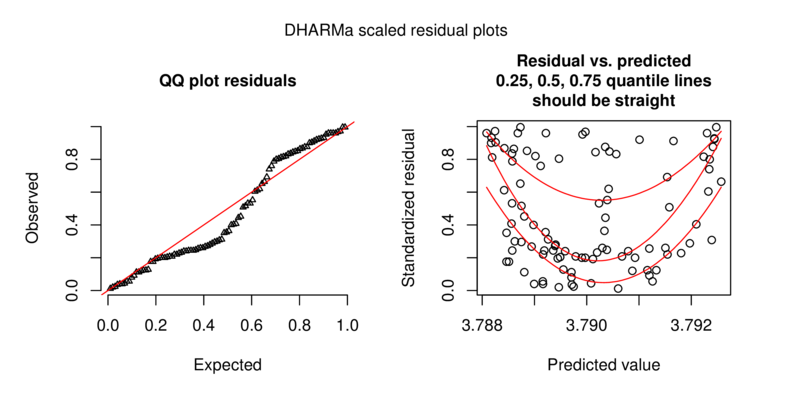
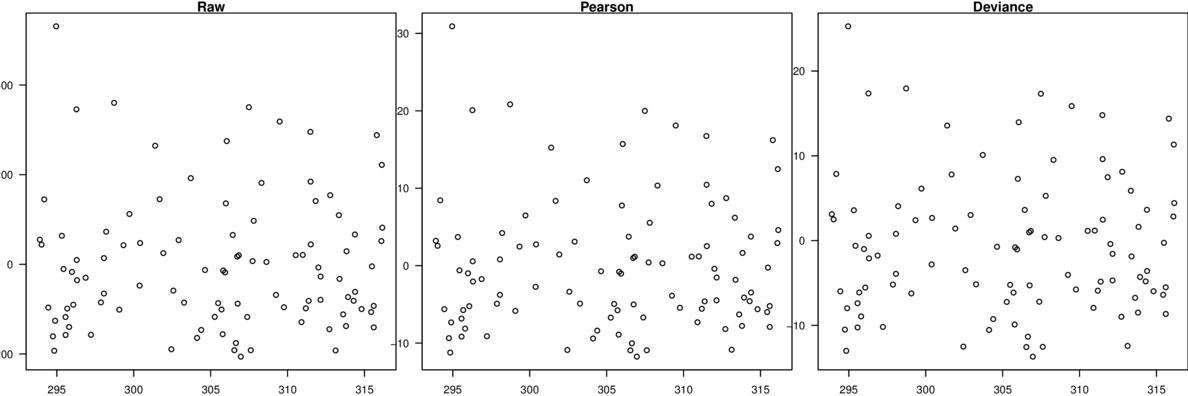

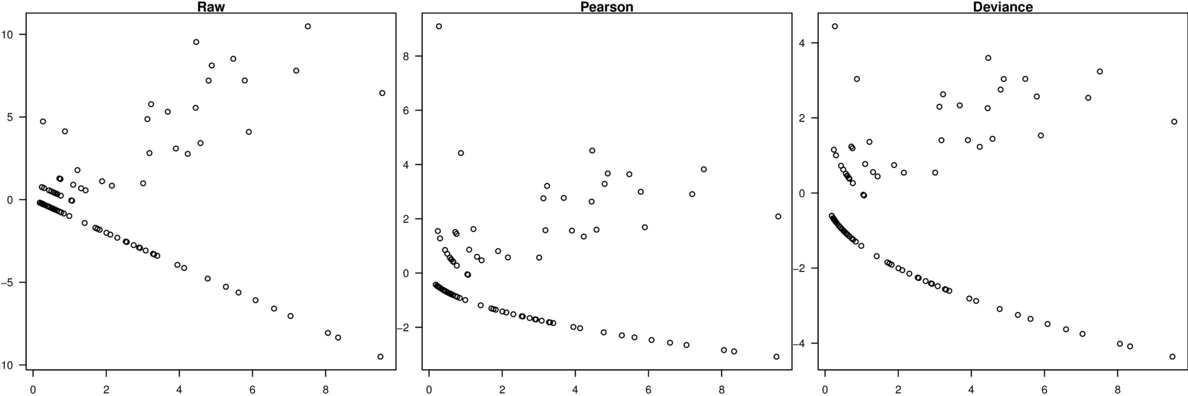
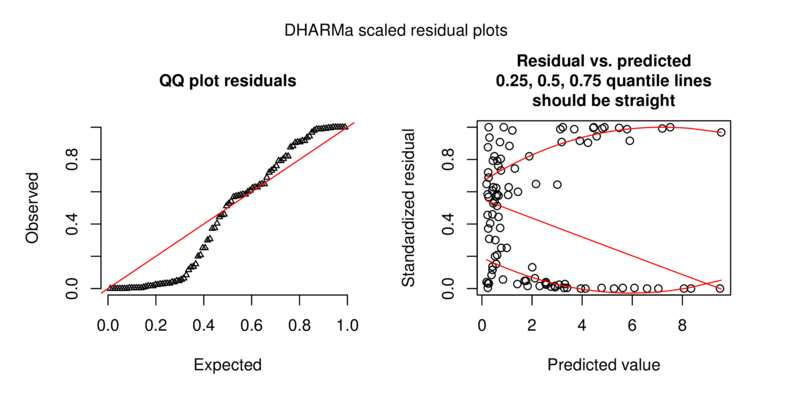
\[p(y_i)=\frac{\Gamma(y_i+\omega)}{\Gamma(\omega)y!}\times\frac{\mu_i^{y_i}\omega^\omega}{(\mu_i+\omega)^{\mu_i+\omega}}\]
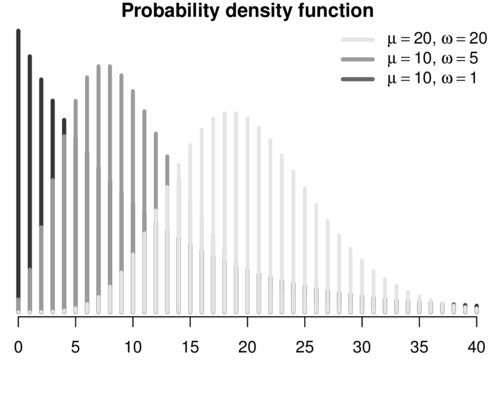
\[p(y_i)=\frac{\Gamma(y_i+\omega)}{\Gamma(\omega)y!}\times\frac{\mu_i^{y_i}\omega^\omega}{(\mu_i+\omega)^{\mu_i+\omega}}\]
| Format of quinn.csv data files | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|

|
SEASON DENSITY RECRUITS SQRTRECRUITS GROUP
1 Spring Low 15 3.872983 SpringLow
2 Spring Low 10 3.162278 SpringLow
3 Spring Low 13 3.605551 SpringLow
4 Spring Low 13 3.605551 SpringLow
5 Spring Low 5 2.236068 SpringLow
6 Spring High 11 3.316625 SpringHigh| Format of moth.csv data files | |||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|

|
||||||||||||||||||||||||||||||||||||
METERS A P HABITAT
1 25 9 8 NWsoak
2 37 3 20 SWsoak
3 109 7 9 Lowerside
4 10 0 2 Lowerside
5 133 9 1 Upperside
6 26 3 18 Disturbed[1] 6.735895e-07[1] 0.175419[1] 0[1] 0.04489437[1] 2.700801[1] 1.228093[1] 8.843367[1] 1.452578[1] 2.93723[1] 5.462792[1] 1.455681[1] 1.371939
FALSE TRUE
0.85 0.15
FALSE TRUE
0.994 0.006 df AICc
moths.glmO 7 312.4946
moths.glmC 8 229.3589
moths.glmNBO 8 233.1423
moths.glmNBC 9 213.2019
Call:
glm.nb(formula = A ~ log(METERS) + HABITAT, data = moths, init.theta = 4.195676174,
link = log)
Deviance Residuals:
Min 1Q Median 3Q Max
-2.4806 -1.2400 -0.0062 0.6398 1.9712
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -0.1076 0.4589 -0.235 0.81455
log(METERS) 0.1451 0.1372 1.057 0.29041
HABITATLowerside 1.2864 0.4710 2.731 0.00631 **
HABITATNEsoak 0.4293 0.5858 0.733 0.46369
HABITATNWsoak 2.8306 0.5392 5.250 1.52e-07 ***
HABITATSEsoak 1.3327 0.5092 2.617 0.00887 **
HABITATSWsoak 1.4899 0.5981 2.491 0.01274 *
HABITATUpperside 1.0764 0.6979 1.542 0.12299
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for Negative Binomial(4.1957) family taken to be 1)
Null deviance: 105.899 on 39 degrees of freedom
Residual deviance: 46.582 on 32 degrees of freedom
AIC: 207.2
Number of Fisher Scoring iterations: 1
Theta: 4.20
Std. Err.: 1.96
2 x log-likelihood: -189.202 HABITAT METER fit lower upper
1 Disturbed 45.5 1.562453 0.5638782 4.329411
2 Lowerside 45.5 5.655624 3.0673703 10.427850
3 NEsoak 45.5 2.400197 1.2093558 4.763650
4 NWsoak 45.5 26.492966 13.6799769 51.306904
5 SEsoak 45.5 5.923526 3.4228572 10.251134
6 SWsoak 45.5 6.931826 3.3129525 14.503741Error in eval(expr, envir, enclos): could not find function "ggplot"