Simple regression trees
- split (partition) data up into major chunks
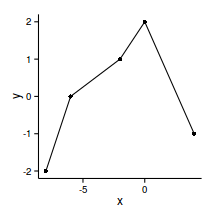
single models fail to capture complexity
Feature complexity
(Multi)collinearity
CART

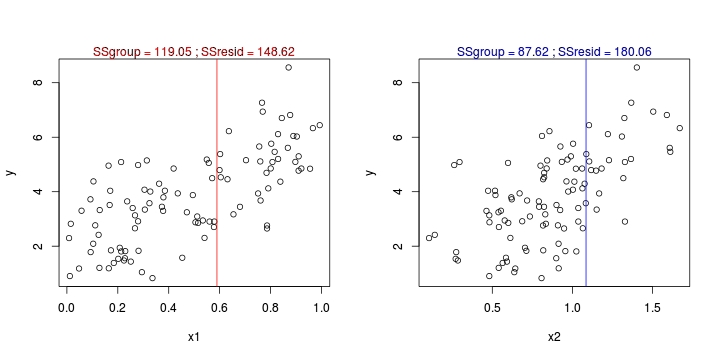
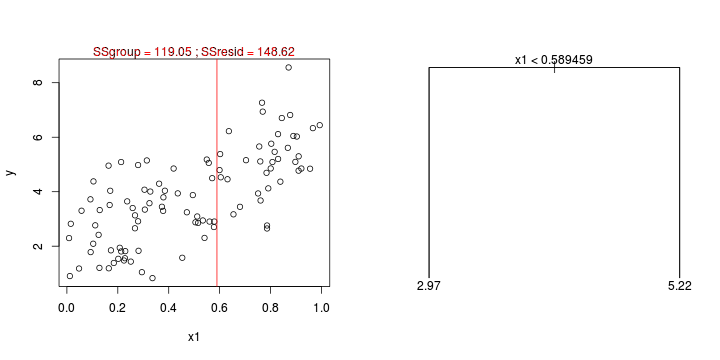
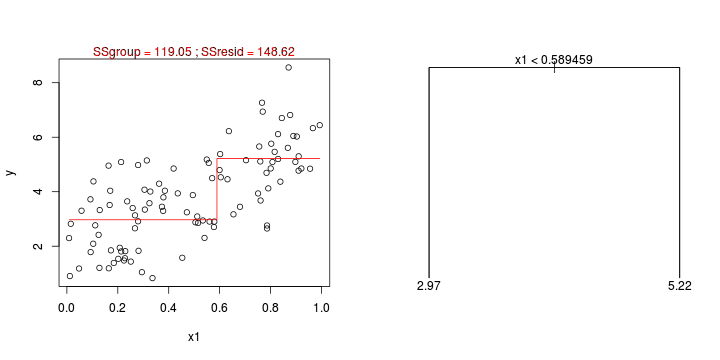
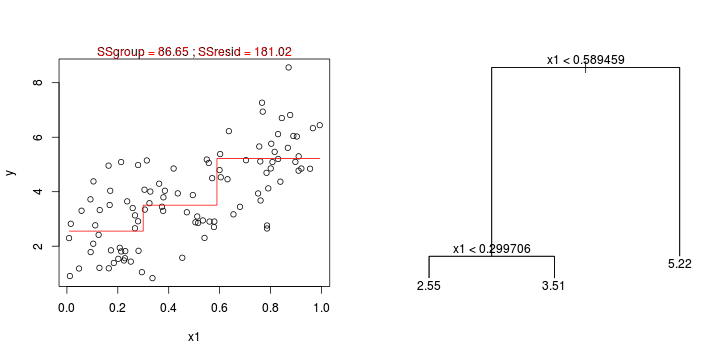

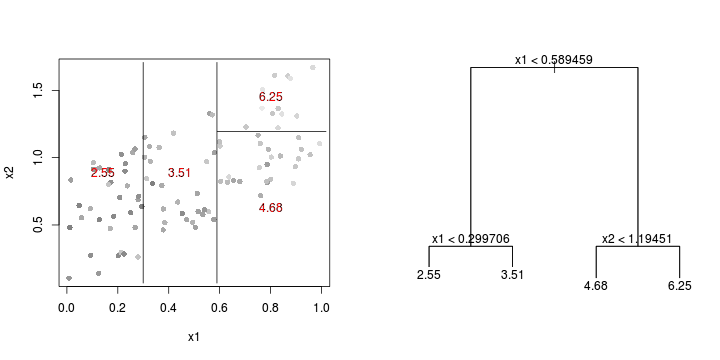
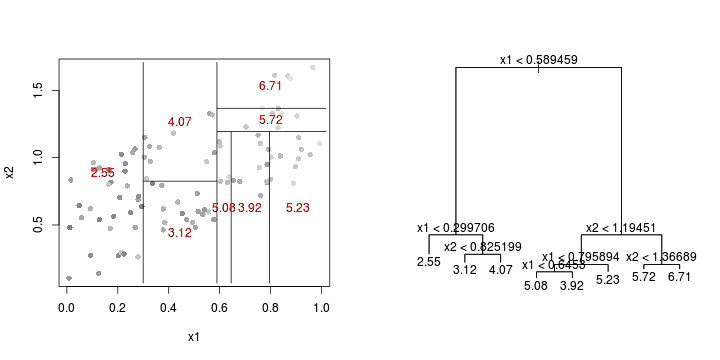

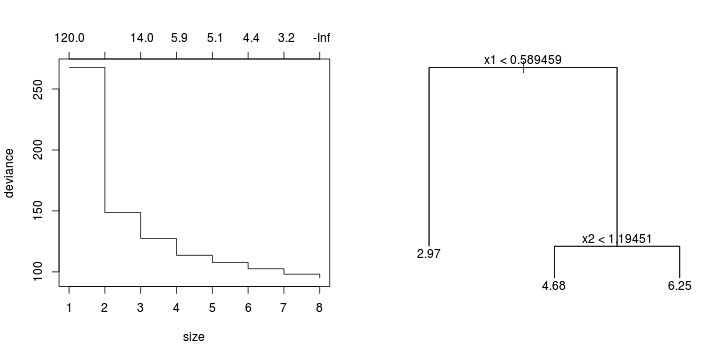
library(tree)library(rpart)Error: unused argument (to = c(0.5, 2.5))Error: unused argument (to = c(0, 1))Error: object 'at' not found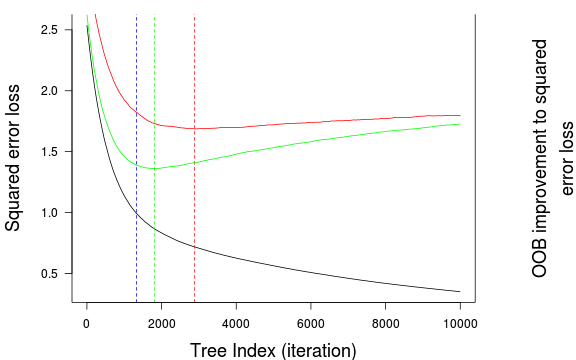

var rel.inf
x1 x1 55.06
x2 x2 44.94[1] 0.5952paruelo <- read.table('../data/paruelo.csv', header=T, sep=',', strip.white=T)
head(paruelo) C3 LAT LONG MAP MAT JJAMAP DJFMAP
1 0.65 46.40 119.55 199 12.4 0.12 0.45
2 0.65 47.32 114.27 469 7.5 0.24 0.29
3 0.76 45.78 110.78 536 7.2 0.24 0.20
4 0.75 43.95 101.87 476 8.2 0.35 0.15
5 0.33 46.90 102.82 484 4.8 0.40 0.14
6 0.03 38.87 99.38 623 12.0 0.40 0.11library(tree)
paruelo.tree <- tree(C3 ~ LAT+LONG+MAP+MAT+JJAMAP,
data=paruelo)
plot(residuals(paruelo.tree)~predict(paruelo.tree))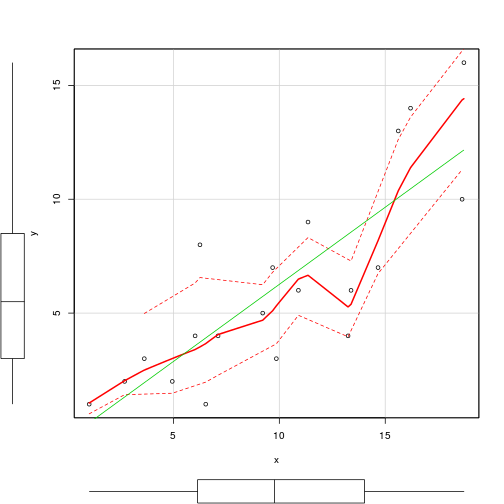
plot(paruelo.tree)
text(paruelo.tree, cex=0.75)
plot(prune.tree(paruelo.tree))
paruelo.tree1 <- prune.tree(paruelo.tree, best=4)
plot(residuals(paruelo.tree1)~predict(paruelo.tree1))
plot(paruelo.tree1)
text(paruelo.tree1)
summary(paruelo.tree1)
Regression tree:
snip.tree(tree = paruelo.tree, nodes = c(7L, 5L, 4L, 6L))
Variables actually used in tree construction:
[1] "LAT" "MAT"
Number of terminal nodes: 4
Residual mean deviance: 0.0304 = 2.1 / 69
Distribution of residuals:
Min. 1st Qu. Median Mean 3rd Qu. Max.
-0.4220 -0.1010 -0.0325 0.0000 0.0787 0.4820 paruelo.tree1$frame var n dev yval splits.cutleft
1 LAT 73 4.9093 0.2714 <42.785
2 MAT 50 1.6364 0.1592 <7.25
4 <leaf> 12 0.4384 0.3425
5 <leaf> 38 0.6674 0.1013
3 MAT 23 1.2762 0.5152 <6.9
6 <leaf> 12 0.6912 0.4083
7 <leaf> 11 0.2984 0.6318
splits.cutright
1 >42.785
2 >7.25
4
5
3 >6.9
6
7 library(scales)
#ys <- with(paruelo,
#rescale(C3, from=c(min(C3), max(C3)),
#to=c(0.8,0)))
#plot(paruelo$LONG,paruelo$LAT,col=grey(ys),
#pch=20,xlab="Longitude",ylab="Latitude")
#partition.tree(paruelo.tree1,ordvars=c("MAT","LAT"),add=TRUE)
#partition.tree(paruelo.tree1,add=TRUE)
#Prediction
xlat <- seq(min(paruelo$LAT), max(paruelo$LAT), l=100)
#xlong <- mean(paruelo$LONG)
#xmat <- mean(paruelo$MAT)
pred <- predict(paruelo.tree1,
newdata=data.frame(LAT=xlat, LONG=mean(paruelo$LONG), MAT=mean(paruelo$MAT),
MAP=mean(paruelo$MAP),
JJAMAP=mean(paruelo$JJAMAP)))
par(mfrow=c(1,2))
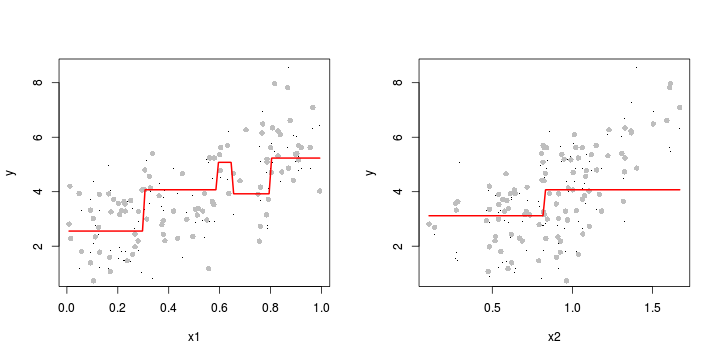
plot(C3~LAT, paruelo, type="p",pch=16, cex=0.2)
points(I(predict(paruelo.tree1)-resid(paruelo.tree1))
~LAT, paruelo, type="p",pch=16, col="grey")
lines(pred~xlat, col="red", lwd=2)
xmat <- seq(min(paruelo$MAT), max(paruelo$MAT), l=100)
#xlat <- mean(paruelo$LAT)
pred <- predict(paruelo.tree1,
newdata=data.frame(LAT=mean(paruelo$LAT), LONG=mean(paruelo$LONG), MAT=xmat,
MAP=mean(paruelo$MAP),
JJAMAP=mean(paruelo$JJAMAP)))
plot(C3~MAT, paruelo, type="p",pch=16, cex=0.2)
points(I(predict(paruelo.tree1)-resid(paruelo.tree1))~MAT, paruelo, type="p",pch=16, col="grey")
lines(pred~xmat, col="red", lwd=2)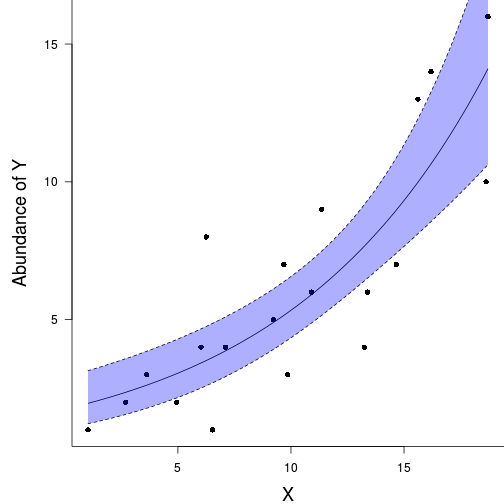
#xlong <- seq(min(paruelo$LONG), max(paruelo$LONG), l=100)
##xlat <- mean(paruelo$LAT)
#pred <- predict(paruelo.tree1,
# newdata=data.frame(LAT=mean(paruelo$LAT), LONG=xlong, MAT=mean(paruelo$MAT),
# MAP=mean(paruelo$MAP),
# JJAMAP=mean(paruelo$JJAMAP)))
#plot(C3~LONG, paruelo, type="p",pch=16, cex=0.2)
#points(I(predict(paruelo.tree1)-resid(paruelo.tree1))~LONG, paruelo, type="p",pch=16, col="grey")
#lines(pred~xlong, col="red", lwd=2)
## paruelo.tree <- tree(C3 ~ LAT+LONG+MAP+MAT+
## JJAMAP+DJFMAP, data=paruelo)
## plot(paruelo.tree)
## text(paruelo.tree, cex=0.75)
## xlat <- seq(min(paruelo$LAT), max(paruelo$LAT), l=100)
## xlong <- mean(paruelo$LONG)
## xmat<- mean(paruelo$MAT)
## xmap<- mean(paruelo$MAP)
## xjjamap<- mean(paruelo$JJAMAP)
## xdjfmap<- mean(paruelo$DJFMAP)
## pp <- predict(paruelo.tree1, newdata=data.frame(LAT=xlat, LONG=xlong, MAT=xmat, MAP=xmap, JJAMAP=xjjamap, DJFMAP=xdjfmap))
## par(mfrow=c(1,2))
## plot(C3~LAT, paruelo, type="p",pch=16, cex=0.2)
## points(I(predict(paruelo.tree1)-resid(paruelo.tree1))~LAT, paruelo, type="p",pch=16, col="grey")
## lines(pp~xlat, col="red", lwd=2)
## xlat <- mean(paruelo$LAT)
## xlong <- mean(paruelo$LONG)
## xmat<- seq(min(paruelo$MAT), max(paruelo$MAT), l=100)
## xmap<- mean(paruelo$MAP)
## xjjamap<- mean(paruelo$JJAMAP)
## xdjfmap<- mean(paruelo$DJFMAP)
## pp <- predict(paruelo.tree1, newdata=data.frame(LAT=xlat, LONG=xlong, MAT=xmat, MAP=xmap, JJAMAP=xjjamap, DJFMAP=xdjfmap))
## par(mfrow=c(1,2))
## plot(C3~MAT, paruelo, type="p",pch=16, cex=0.2)
## points(I(predict(paruelo.tree1)-resid(paruelo.tree1))~MAT, paruelo, type="p",pch=16, col="grey")
## lines(pp~xmat, col="red", lwd=2)
## Now GBM
library(gbm)
paruelo.gbm <- gbm(C3~LAT+LONG+MAP+MAT+JJAMAP+DJFMAP,
data=paruelo,
distribution="gaussian",
n.trees=10000,
interaction.depth=3, # 1: additive model, 2: two-way interactions, etc.
cv.folds=3,
train.fraction=0.75,
bag.fraction=0.5,
shrinkage=0.001,
n.minobsinnode=2)
## Out of Bag method of determining number of iterations
(best.iter <- gbm.perf(paruelo.gbm, method="test"))[1] 1533(best.iter <- gbm.perf(paruelo.gbm, method="OOB"))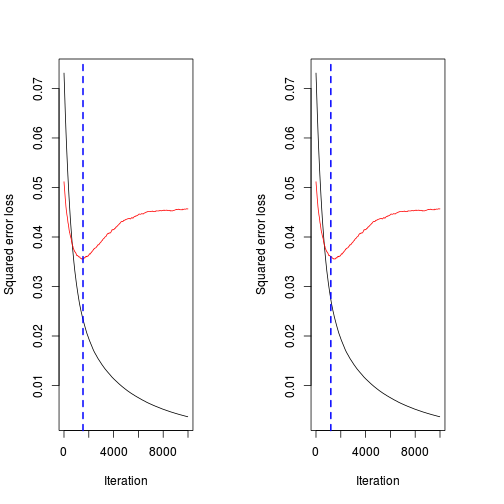
[1] 1197(best.iter <- gbm.perf(paruelo.gbm, method="OOB",oobag.curve=TRUE,overlay=TRUE, plot.it=TRUE))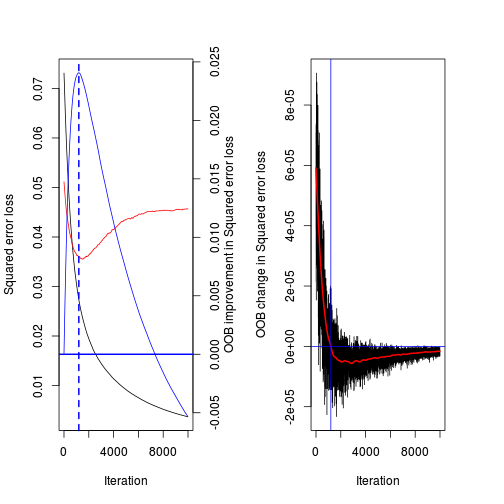
[1] 1197(best.iter <- gbm.perf(paruelo.gbm, method="cv"))[1] 1844par(mfrow=c(1,2))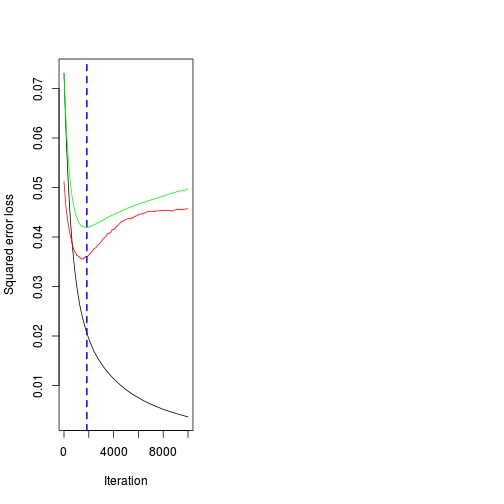
best.iter <- gbm.perf(paruelo.gbm, method="cv",
oobag.curve=TRUE,overlay=TRUE,plot.it=TRUE)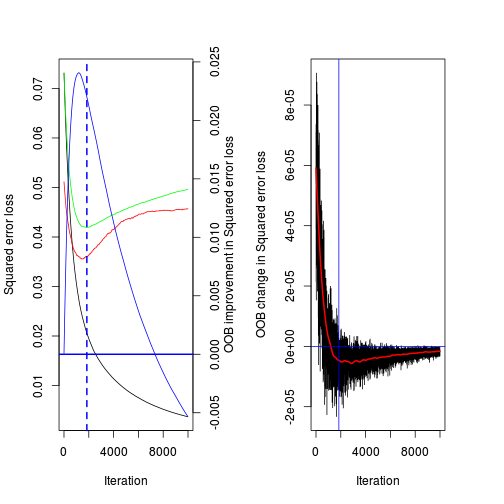
best.iter[1] 1844par(mfrow=c(3,2),mar=c(4,5,0,0))
plot(paruelo.gbm, 1, n.tree=best.iter)
plot(paruelo.gbm, 2, n.tree=best.iter)
plot(paruelo.gbm, 3, n.tree=best.iter)
plot(paruelo.gbm, 4, n.tree=best.iter)
plot(paruelo.gbm, 5, n.tree=best.iter)
plot(paruelo.gbm, 6, n.tree=best.iter)
par(mfrow=c(3,2),mar=c(4,5,0,0))
plot(paruelo.gbm, 1, n.tree=best.iter, ylim=c(0.1,0.6))
plot(paruelo.gbm, 2, n.tree=best.iter, ylim=c(0.1,0.6))
plot(paruelo.gbm, 3, n.tree=best.iter, ylim=c(0.1,0.6))
plot(paruelo.gbm, 4, n.tree=best.iter, ylim=c(0.1,0.6))
plot(paruelo.gbm, 5, n.tree=best.iter, ylim=c(0.1,0.6))
plot(paruelo.gbm, 6, n.tree=best.iter, ylim=c(0.1,0.6))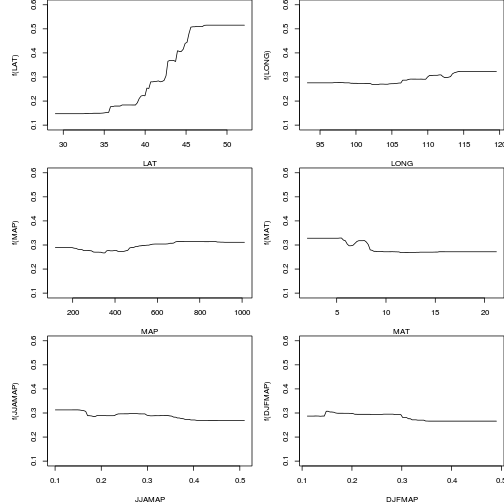
pp<-plot(paruelo.gbm, c(2,1), best.iter,
return.grid=TRUE)
#scatter3d(x=pp$LONG, y=pp$LAT, z=pp$y)
par(mfrow=c(1,1))
library(scatterplot3d)
scatterplot3d(x=pp$LONG, y=pp$LAT, z=pp$y, angle=24)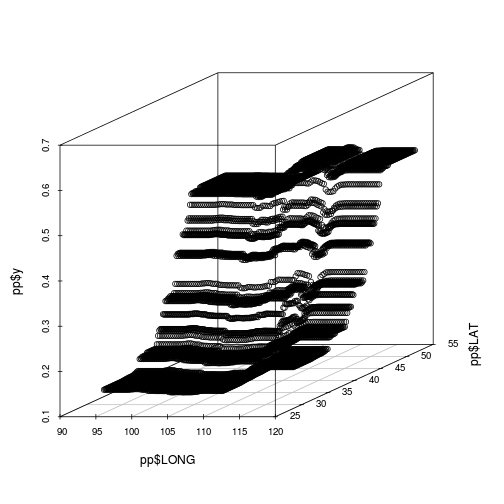
#with(pp[1:20,],persp(x=LONG, y=LAT, z=y))
xyz <- with(pp,unique(cbind(x=LONG,y=LAT,z=pp$y)))
persp(xyz, theta=-60, phi=-10)
par(mfrow=c(1,1))
summary(paruelo.gbm)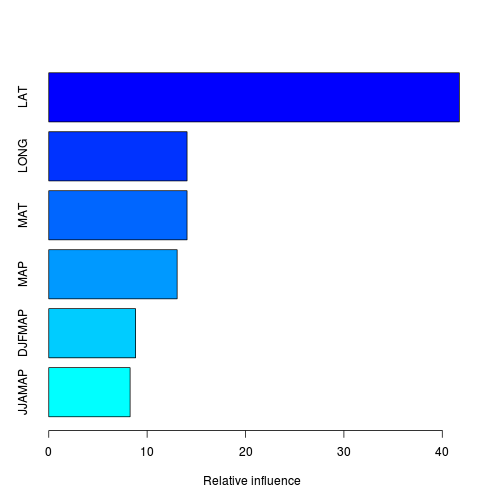
var rel.inf
LAT LAT 41.736
LONG LONG 14.058
MAT MAT 14.057
MAP MAP 13.052
DJFMAP DJFMAP 8.827
JJAMAP JJAMAP 8.270## interact.gbm’ computes Friedman's H-statistic to assess the
## relative strength of interaction effects in non-linear models. H
## is on the scale of [0-1] with higher values indicating larger
## interaction effects. To connect to a more familiar measure, if x_1
## and x_2 are uncorrelated covariates with mean 0 and variance 1 and
## the model is of the form
## y=beta_0+beta_1x_1+beta_2x_2+beta_3x_3
## then
## H=\frac{beta_3}{sqrt{beta_1^2+beta_2^2+beta_3^2}}
## Note that if the main effects are weak, the estimated H will be
## unstable. For example, if (in the case of a two-way interaction)
## neither main effect is in the selected model (relative influence
## is zero), the result will be 0/0. Also, with weak main effects,
## rounding errors can result in values of H > 1 which are not
## possible.
interact.gbm(paruelo.gbm, paruelo,1:2,n.tree=best.iter)[1] 0.06536interact.gbm(paruelo.gbm, paruelo,c(1,3),n.tree=best.iter)[1] 0.0537interact.gbm(paruelo.gbm, paruelo,c(1,2,3),n.tree=best.iter)[1] 0.002928## terms <- attr(paruelo.gbm$Terms,"term.labels")
## for (i in 1:5) {
## for (j in (i+1):6) {
## print(paste(terms[i],":",terms[j],"=",
## interact.gbm(paruelo.gbm, paruelo,c(i,j),n.tree=best.iter),sep=""))
## }
## }
terms <- attr(paruelo.gbm$Terms,"term.labels")
paruelo.int <- NULL
for (i in 1:(length(terms)-1)) {
for (j in (i+1):length(terms)) {
paruelo.int <- rbind(paruelo.int,
data.frame(Var1=terms[i], Var2=terms[j],
"H.stat"=interact.gbm(paruelo.gbm, paruelo,c(i,j),
n.tree=best.iter)
))
}
}
paruelo.int Var1 Var2 H.stat
1 LAT LONG 0.06536
2 LAT MAP 0.05370
3 LAT MAT 0.09307
4 LAT JJAMAP 0.04462
5 LAT DJFMAP 0.04217
6 LONG MAP 0.04100
7 LONG MAT 0.03779
8 LONG JJAMAP 0.02226
9 LONG DJFMAP 0.09464
10 MAP MAT 0.09162
11 MAP JJAMAP 0.02688
12 MAP DJFMAP 0.01857
13 MAT JJAMAP 0.14502
14 MAT DJFMAP 0.15350
15 JJAMAP DJFMAP 0.08397plot(paruelo.gbm, c(2,1), n.tree=best.iter)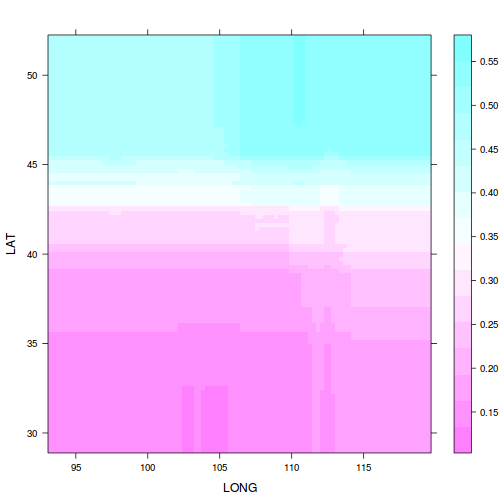
loyn <- read.table('../data/loyn.csv', header=T, sep=',', strip.white=T)
head(loyn) ABUND AREA YR.ISOL DIST LDIST GRAZE ALT
1 5.3 0.1 1968 39 39 2 160
2 2.0 0.5 1920 234 234 5 60
3 1.5 0.5 1900 104 311 5 140
4 17.1 1.0 1966 66 66 3 160
5 13.8 1.0 1918 246 246 5 140
6 14.1 1.0 1965 234 285 3 130loyn$fGRAZE<- factor(loyn$GRAZE)
head(loyn) ABUND AREA YR.ISOL DIST LDIST GRAZE ALT fGRAZE
1 5.3 0.1 1968 39 39 2 160 2
2 2.0 0.5 1920 234 234 5 60 5
3 1.5 0.5 1900 104 311 5 140 5
4 17.1 1.0 1966 66 66 3 160 3
5 13.8 1.0 1918 246 246 5 140 5
6 14.1 1.0 1965 234 285 3 130 3library(tree)
loyn.tree <- tree(ABUND~AREA+YR.ISOL+DIST+
LDIST+fGRAZE+ALT, data=loyn, mindev=0)
plot(residuals(loyn.tree)~predict(loyn.tree))
plot(loyn.tree, type="uniform")
text(loyn.tree,cex=1, all=T)
text(loyn.tree,lab=paste("n"),
cex=1,adj=c(0,2), splits=F)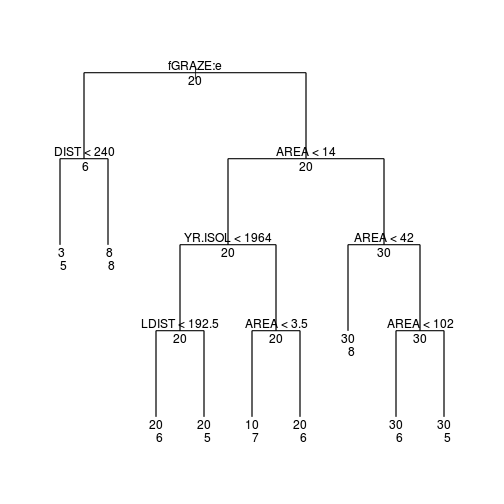
plot(prune.tree(loyn.tree))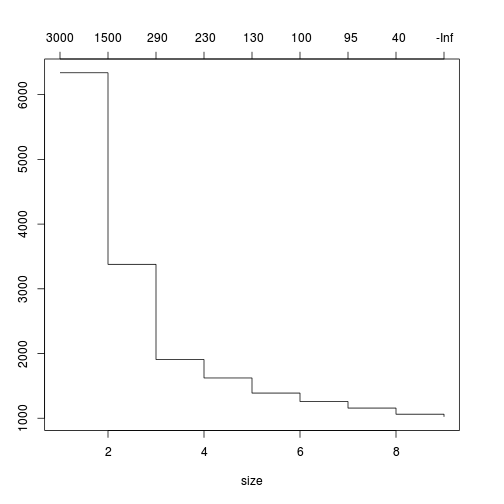
loyn.tree.prune<-prune.tree(loyn.tree,
best=3)
plot(loyn.tree.prune, type="uniform")
text(loyn.tree.prune,cex=1, all=T)
text(loyn.tree.prune,lab=paste("n"),
cex=1,adj=c(0,2), splits=F)
library(rpart)
loyn.rpart<-rpart(ABUND~AREA+YR.ISOL+
DIST+LDIST+GRAZE+ALT, data=loyn)
plot(loyn.rpart)
text(loyn.rpart,use.n=TRUE)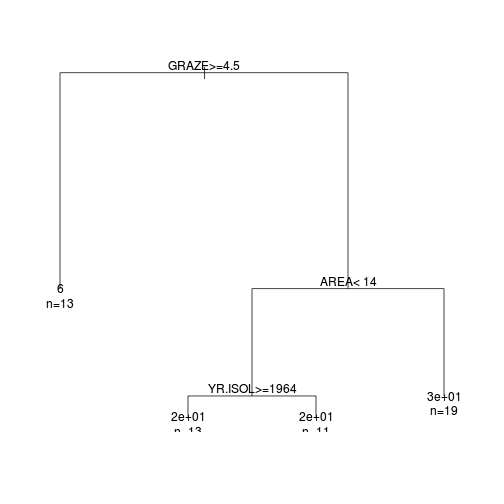
#gbm
library(gbm)
loyn.gbm <- gbm(ABUND~DIST+LDIST+AREA+
fGRAZE+ALT+YR.ISOL,
data=loyn,
distribution="gaussian",
train=1,
interaction.depth=3,
cv.folds=1,
shrinkage=0.001,
bag.fraction=0.5,
n.minobsinnode=2,
n.trees=10000)Error: object 'p' not found#loyn.gbm <- gbm(C3~LAT + LONG + MAP + MAT + JJAMAP + DJFMAP, data=loyn,
# interaction.depth=3, cv.folds=3, dist="bernoulli")
#best.iter <- gbm.perf(loyn.gbm, method="test") #see graph
#best.iter <- gbm.perf(loyn.gbm, method="OOB", plot.it=TRUE) #see graph
par(mfrow=c(1,2))
(best.iter <- gbm.perf(loyn.gbm, method="OOB",
overlay=TRUE,oobag.curve=TRUE))Error: object 'loyn.gbm' not found#print(best.iter)
summary(loyn.gbm,n.trees=best.iter) #see graphError: object 'loyn.gbm' not foundpar(mfrow=c(2,3))
plot(loyn.gbm, 1, best.iter)Error: object 'loyn.gbm' not foundplot(loyn.gbm, 2, best.iter)Error: object 'loyn.gbm' not foundplot(loyn.gbm, 3, best.iter)Error: object 'loyn.gbm' not foundplot(loyn.gbm, 4, best.iter)Error: object 'loyn.gbm' not foundplot(loyn.gbm, 5, best.iter)Error: object 'loyn.gbm' not foundplot(loyn.gbm, 6, best.iter)Error: object 'loyn.gbm' not foundpar(mfrow=c(2,3))
plot(loyn.gbm, 1, best.iter, ylim=c(10,30))Error: object 'loyn.gbm' not foundplot(loyn.gbm, 2, best.iter, ylim=c(10,30))Error: object 'loyn.gbm' not foundplot(loyn.gbm, 3, best.iter, ylim=c(10,30))Error: object 'loyn.gbm' not foundplot(loyn.gbm, 4, best.iter, ylim=c(10,30))Error: object 'loyn.gbm' not foundplot(loyn.gbm, 5, best.iter, ylim=c(10,30))Error: object 'loyn.gbm' not foundplot(loyn.gbm, 6, best.iter, ylim=c(10,30))Error: object 'loyn.gbm' not founddists <- seq(min(loyn$DIST),max(loyn$DIST),l=100)
ldists <- seq(min(loyn$LDIST),max(loyn$LDIST),l=100)
areas <- seq(min(loyn$AREA),max(loyn$AREA),l=100)
grazes <- seq(min(loyn$GRAZE),max(loyn$GRAZE),l=100)
alts <- seq(min(loyn$ALT),max(loyn$ALT),l=100)
yrs <- seq(min(loyn$YR.ISOL),max(loyn$YR.ISOL),l=100)
new<-predict(loyn.gbm,
newdata=data.frame(
DIST=10,
LDIST=10,
AREA=500,
fGRAZE=2,
ALT=200,
YR.ISOL=1960
), n.trees=best.iter
)Error: object 'loyn.gbm' not foundnewfunction (Class, ...)
{
ClassDef <- getClass(Class, where = topenv(parent.frame()))
value <- .Call(C_new_object, ClassDef)
initialize(value, ...)
}
<bytecode: 0x3239690>
<environment: namespace:methods>#plot(aa~areas, type="l",ylim=c(7,30))
aa<-predict(loyn.gbm, newdata=data.frame(
DIST=800,
LDIST=800,
AREA=areas,
fGRAZE=1,
ALT=1000,
YR.ISOL=10
), n.trees=best.iter
)Error: object 'loyn.gbm' not foundplot(aa~areas, type="l")Error: object 'aa' not foundplot(aa~areas, type="l",log="x")Error: object 'aa' not foundaa<-predict(loyn.gbm, newdata=data.frame(
DIST=mean(dists),
LDIST=mean(ldists),
AREA=areas,
GRAZE=mean(grazes),
ALT=200,
YR.ISOL=mean(yrs)
), n.trees=best.iter
)Error: object 'loyn.gbm' not foundaa<-predict(loyn.gbm, newdata=data.frame(
DIST=mean(loyn$DIST),
LDIST=mean(loyn$LDIST),
AREA=mean(loyn$AREA),
GRAZE=grazes,
ALT=mean(loyn$ALT),
YR.ISOL=mean(loyn$YR.ISOL)
), n.trees=best.iter
)Error: object 'loyn.gbm' not foundplot(aa~grazes, type="l")Error: object 'aa' not foundterms <- attr(loyn.gbm$Terms,"term.labels")Error: object 'loyn.gbm' not foundloyn.int <- NULL
for (i in 1:(length(terms)-1)) {
for (j in (i+1):length(terms)) {
loyn.int <- rbind(loyn.int,
data.frame(Var1=terms[i], Var2=terms[j],
"H.stat"=interact.gbm(loyn.gbm, loyn,c(i,j),n.tree=best.iter)
))
}
}Error: object 'loyn.gbm' not foundloyn.intNULLloyn.gbm <- gbm(ABUND~DIST+LDIST+AREA+
factor(GRAZE)+ALT+YR.ISOL, data=loyn,
interaction.depth=3, cv.folds=3,train=0.5,
n.minobsinnode=2, n.trees=10000)Distribution not specified, assuming gaussian ...Error: subscript out of boundsbest.iter <- gbm.perf(loyn.gbm, method="cv") #see graphError: object 'loyn.gbm' not foundprint(best.iter)[1] 1844summary(loyn.gbm,n.trees=best.iter) #see graphError: object 'loyn.gbm' not foundplot(loyn.gbm, 3:4, best.iter, ylim=c(10,30))Error: object 'loyn.gbm' not found##PARUELO
paruelo <- read.table('../data/paruelo.csv', header=T, sep=',', strip.white=T)
head(paruelo) C3 LAT LONG MAP MAT JJAMAP DJFMAP
1 0.65 46.40 119.55 199 12.4 0.12 0.45
2 0.65 47.32 114.27 469 7.5 0.24 0.29
3 0.76 45.78 110.78 536 7.2 0.24 0.20
4 0.75 43.95 101.87 476 8.2 0.35 0.15
5 0.33 46.90 102.82 484 4.8 0.40 0.14
6 0.03 38.87 99.38 623 12.0 0.40 0.11## @knitr ws4-Q1-2
#via car's scatterplotMatrix function
library(car)
scatterplotMatrix(~C3+LAT+LONG+MAP+MAT+JJAMAP+DJFMAP, data=paruelo, diagonal='boxplot')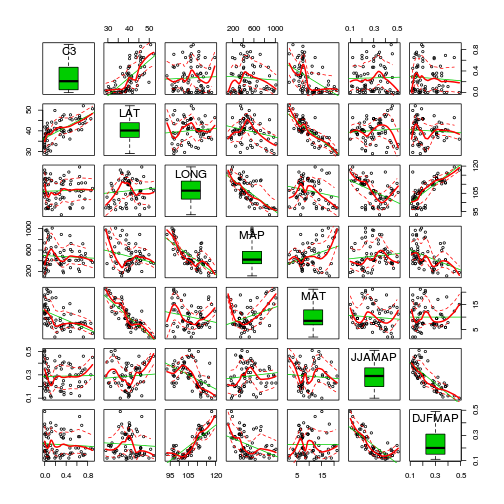
scatterplotMatrix(~sqrt(C3)+LAT+LONG+MAP+MAT+JJAMAP+log10(DJFMAP), data=paruelo, diagonal='boxplot')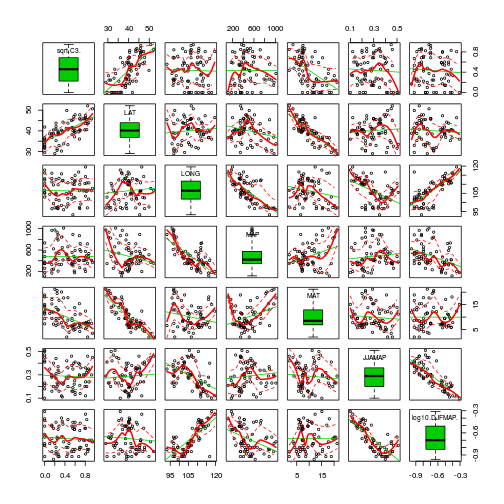
library(car)
vif(lm(sqrt(paruelo$C3) ~ LAT + LONG + MAP + MAT + JJAMAP + DJFMAP, data=paruelo)) LAT LONG MAP MAT JJAMAP DJFMAP
3.503 5.268 2.799 3.743 3.163 5.710 library(car)
1/vif(lm(sqrt(paruelo$C3) ~ LAT + LONG + MAP + MAT + JJAMAP + log10(DJFMAP), data=paruelo)) LAT LONG MAP
0.2809 0.2005 0.3579
MAT JJAMAP log10(DJFMAP)
0.2665 0.3130 0.1829 paruelo$cLAT <- scale(paruelo$LAT, scale=F)
paruelo$cLONG <- scale(paruelo$LONG, scale=F)
## @knitr Q1-5
paruelo.lm <- lm(sqrt(C3) ~ cLAT*cLONG, data=paruelo)
summary(paruelo.lm)
Call:
lm(formula = sqrt(C3) ~ cLAT * cLONG, data = paruelo)
Residuals:
Min 1Q Median 3Q Max
-0.5131 -0.1343 -0.0113 0.1409 0.3894
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.428266 0.023435 18.27 < 2e-16
cLAT 0.043694 0.004867 8.98 3.3e-13
cLONG -0.002877 0.003684 -0.78 0.4375
cLAT:cLONG 0.002282 0.000747 3.06 0.0032
(Intercept) ***
cLAT ***
cLONG
cLAT:cLONG **
---
Signif. codes:
0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.199 on 69 degrees of freedom
Multiple R-squared: 0.54, Adjusted R-squared: 0.52
F-statistic: 27 on 3 and 69 DF, p-value: 1.13e-11## @knitr Q1-6
#via the car package
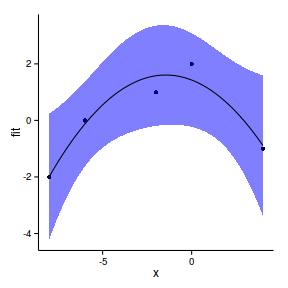
avPlots(paruelo.lm, ask=F, indentify.points=F)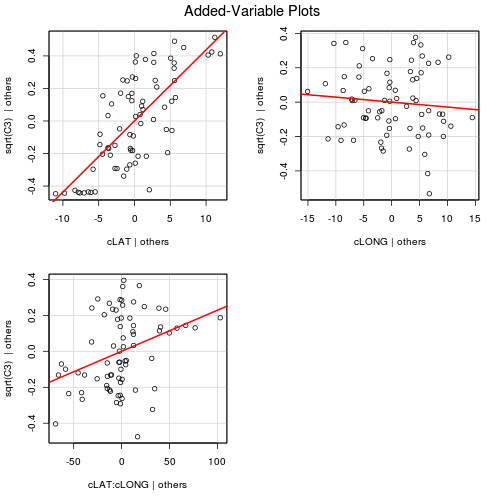
library(effects)
plot(allEffects(paruelo.lm))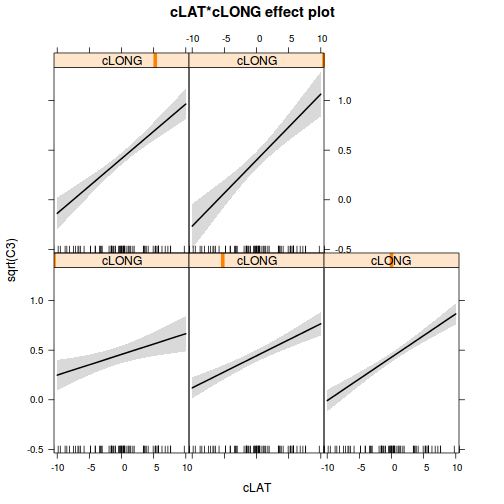
library(tree)
paruelo.tree <- tree(C3~LAT+LONG+MAP+MAT+
JJAMAP+DJFMAP, data=paruelo, mindev=0)
plot(residuals(paruelo.tree)~predict(paruelo.tree))
plot(paruelo.tree, type="uniform")
text(paruelo.tree,cex=1, all=T)
text(paruelo.tree,lab=paste("n"),cex=1,adj=c(0,2), splits=F)
plot(prune.tree(paruelo.tree))
paruelo.tree.prune<-prune.tree(paruelo.tree, best=4)
plot(paruelo.tree.prune, type="uniform")
text(paruelo.tree.prune,cex=1, all=T)
text(paruelo.tree.prune,lab=paste("n"),cex=1,adj=c(0,2), splits=F)
library(gbm)
paruelo.gbm <- gbm(C3~LAT+LONG+MAP+MAT+JJAMAP+DJFMAP,
data=paruelo,
interaction.depth=3,
train=0.5,
cv.folds=3,
n.minobsinnode=2,
n.trees=10000)Distribution not specified, assuming gaussian ...paruelo.gbm <- gbm(C3~LAT+LONG+MAP+MAT+JJAMAP+DJFMAP,
data=paruelo,
interaction.depth=7,
train=0.5,
cv.folds=3,
n.minobsinnode=2,
n.trees=10000)Distribution not specified, assuming gaussian ...#paruelo.gbm <- gbm(C3~LAT + LONG + MAP + MAT + JJAMAP + DJFMAP, data=paruelo,
# interaction.depth=3, cv.folds=3, dist="bernoulli")
best.iter <- gbm.perf(paruelo.gbm,
method="cv")
print(best.iter)[1] 1035summary(paruelo.gbm,n.trees=best.iter) #see graph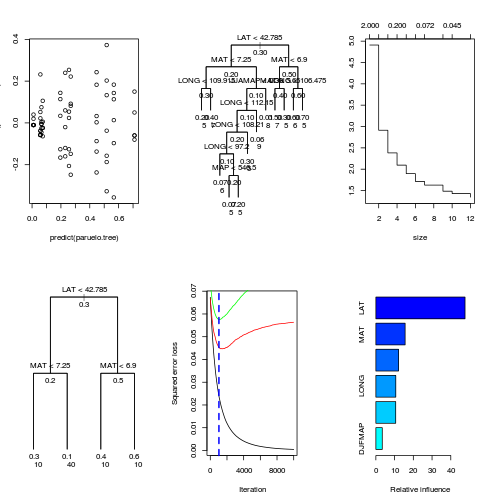
var rel.inf
LAT LAT 47.607
MAT MAT 15.630
MAP MAP 12.095
LONG LONG 10.642
JJAMAP JJAMAP 10.597
DJFMAP DJFMAP 3.429par(mfrow=c(2,3))
plot(paruelo.gbm, 1, best.iter)
plot(paruelo.gbm, 2, best.iter)
plot(paruelo.gbm, 3, best.iter)
plot(paruelo.gbm, 4, best.iter)
plot(paruelo.gbm, 5, best.iter)
plot(paruelo.gbm, 6, best.iter)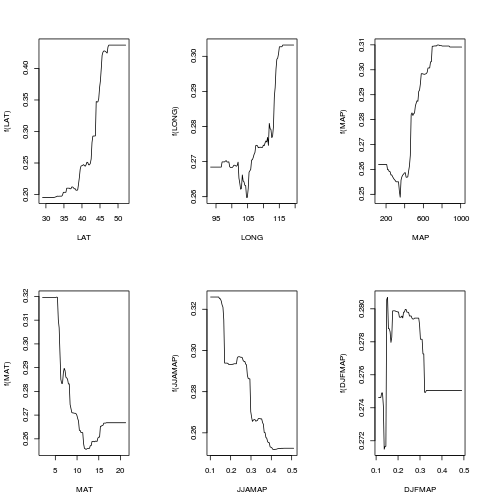
plot(paruelo.gbm, c(2,1), best.iter)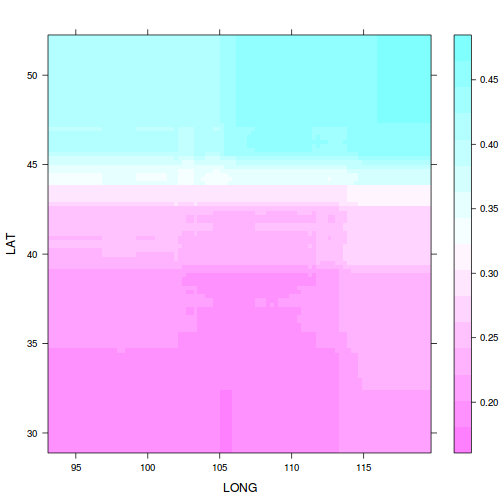
pp<-plot(paruelo.gbm, c(2,1), best.iter,
return.grid=TRUE)
library(ggplot2)
ggplot(pp, aes(y=LAT, x=LONG, fill=y))+
geom_tile()
#scatter3d(x=pp$LONG, y=pp$LAT, z=pp$y)
library(scatterplot3d)
scatterplot3d(x=pp$LONG, y=pp$LAT, z=pp$y, angle=24)
with(pp[1:20,],persp(x=LONG, y=LAT, z=y))Error: increasing 'x' and 'y' values expectedxyz <- with(pp,unique(cbind(x=LONG,y=LAT,z=y)))
persp(xyz, theta=-60, phi=-10)
## tree.screens()
## plot(paruelo.tree)
## text(paruelo.tree)
## tile.tree(paruelo.tree, paruelo$LAT)
## #tile.tree(paruelo.tree, paruelo$LONG)
## close.screen(all = TRUE)
## paruelo.tree1<-prune.tree(paruelo.tree)
## summary(paruelo.tree1)
## paruelo.tree1$frame
## paruelo.tree1<-prune.tree(paruelo.tree,best=2)
## summary(paruelo.tree1)
## paruelo.tree1$frame
## C3.deciles = quantile(paruelo$C3,1:10/10)
## C3.cut = cut(paruelo$C3,breaks=C3.deciles,include.lowest=TRUE)
## plot(paruelo$LONG,paruelo$LAT,col=grey(10:2/11)[C3.cut],pch=20,xlab="Longitude",ylab="Latitude")
## plot(paruelo$LONG,paruelo$LAT,col=grey(10:2/11)[C3.cut],pch=20,xlab="Longitude",ylab="Latitude")
## paruelo.tree1<-prune.tree(paruelo.tree,best=2)
## plot(paruelo$LAT,paruelo$C3,col=grey(10:2/11)[C3.cut],pch=20,xlab="Longitude",ylab="Latitude")
## partition.tree(paruelo.tree1,ordvars=c("LAT"),add=TRUE)
## partition.tree(paruelo.tree1,ordvars=c("LONG","LAT"),add=FALSE)
## plot(paruelo.tree1)
## text(paruelo.tree1, cex=0.75)
## plot(paruelo$C3~paruelo$LAT,col=grey(10:2/11)[C3.cut],pch=20)
## partition.tree(paruelo.tree,ordvars=c("LONG","LAT"),add=TRUE)