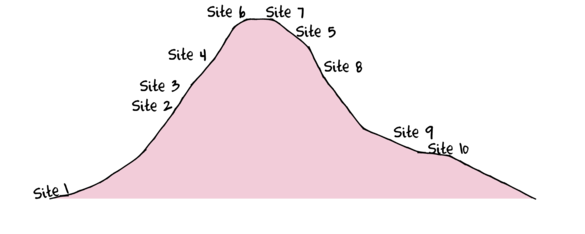
Workshop 15: Q-mode MVA
Murray Logan
06 Aug 2016
R-mode analyses
preserve euclidean (PCA/RDA) or \(\chi^2\) (CA/CCA) distances
based on either correlation or covariance of variables
data restricted by assumptions
Q-mode analyses
based on object similarity
unrestricted choice of similarity/distance matrix
resulting scores not independent
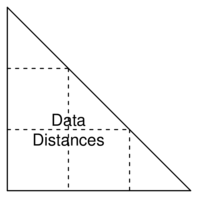
Dissimilarity
Sp1
Sp2
Sp3
Sp4
Site1
2
0
0
5
Site2
13
7
10
5
Site3
9
5
55
93
Site4
10
6
76
81
Site5
0
2
6
0
Distance measures
\[
d_{jk} = \sqrt{\sum{(y_{ji}-y_{ki})^2}}
\]
> library (vegan)
> vegdist (Y,method= "euclidean" ) Site1 Site2 Site3 Site4
Site2 16.431677
Site3 104.129727 98.939375
Site4 107.944430 100.707497 24.228083
Site5 8.306624 15.329710 105.546198 107.596468
Distance measures
\[
\begin{align}
d_{jk} &= \frac{\sum{\mid{}y_{ji}-y_{ki}\mid{}}}{\sum{y_{ji}+y_{ki}}} \\
&= 1-\frac{2\sum{min(y_{ji},y_{ki})}}{\sum{y_{ji}+y_{ki}}}
\end{align}
\]
> library (vegan)
> vegdist (Y,method= "bray" ) Site1 Site2 Site3 Site4
Site2 0.6666667
Site3 0.9171598 0.7055838
Site4 0.9222222 0.7019231 0.1044776
Site5 1.0000000 0.6279070 0.9058824 0.9116022
Distance measures
\[
d_{jk} = \sqrt{\sum{\left(\sqrt{\frac{y_{ji}}{\sum{y_j}}} - \sqrt{\frac{y_{ki}}{\sum{y_k}}}\right)^2}}
\]
> library (vegan)
> dist (decostand (Y,method= "hellinger" )) Site1 Site2 Site3 Site4
Site2 0.8423744
Site3 0.6836053 0.5999300
Site4 0.7657483 0.5608590 0.1092925
Site5 1.4142136 0.7918120 0.9028295 0.8159425
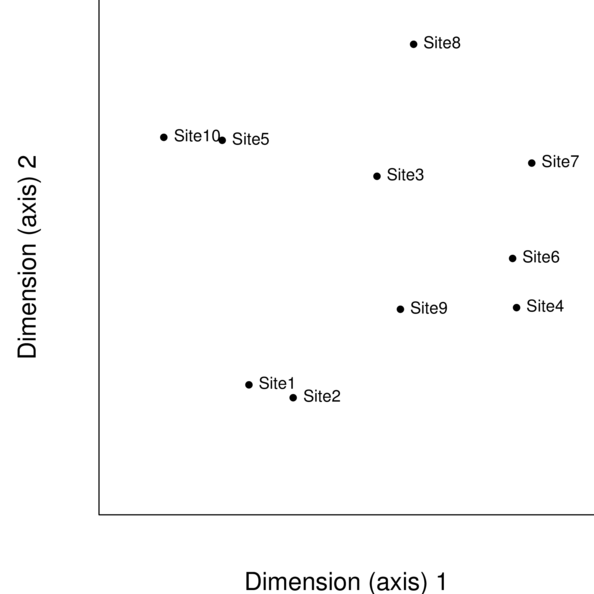
Multidimensional scaling
re-project objects (sites) in reduced dimensional space
must nominate the number of dimensions up-front
optimized patterns for the nominated dimensions
Q-mode analyses
Sites
Sp1
Sp2
Sp3
Sp4
Sp5
Sp6
Sp7
Sp8
Sp9
Sp10
Site1
Site1
5
0
0
65
5
0
0
0
0
0
Site2
Site2
0
0
0
25
39
0
6
23
0
0
Site3
Site3
0
0
0
6
42
0
6
31
0
0
Site4
Site4
0
0
0
0
0
0
0
40
0
14
Site5
Site5
0
0
6
0
0
0
0
34
18
12
Site6
Site6
0
29
12
0
0
0
0
0
22
0
Site7
Site7
0
0
21
0
0
5
0
0
20
0
Site8
Site8
0
0
0
0
13
0
6
37
0
0
Site9
Site9
0
0
0
60
47
0
4
0
0
0
Site10
Site10
0
0
0
72
34
0
0
0
0
0
Multidimensional scaling
generate distance matrix
> data.dist <- vegdist (data[,-1 ], "bray" )
> data.dist Site1 Site2 Site3 Site4 Site5 Site6 Site7 Site8 Site9
Site2 0.6428571
Site3 0.8625000 0.1685393
Site4 1.0000000 0.6870748 0.5539568
Site5 1.0000000 0.7177914 0.6000000 0.2580645
Site6 1.0000000 1.0000000 1.0000000 1.0000000 0.6390977
Site7 1.0000000 1.0000000 1.0000000 1.0000000 0.5862069 0.4128440
Site8 0.9236641 0.4362416 0.2907801 0.3272727 0.4603175 1.0000000 1.0000000
Site9 0.3010753 0.3333333 0.4693878 1.0000000 1.0000000 1.0000000 1.0000000 0.7964072
Site10 0.2265193 0.4070352 0.5811518 1.0000000 1.0000000 1.0000000 1.0000000 0.8395062 0.1336406
Multidimensional scaling
choose # dimensions (k=2)
Multidimensional scaling
random configuration
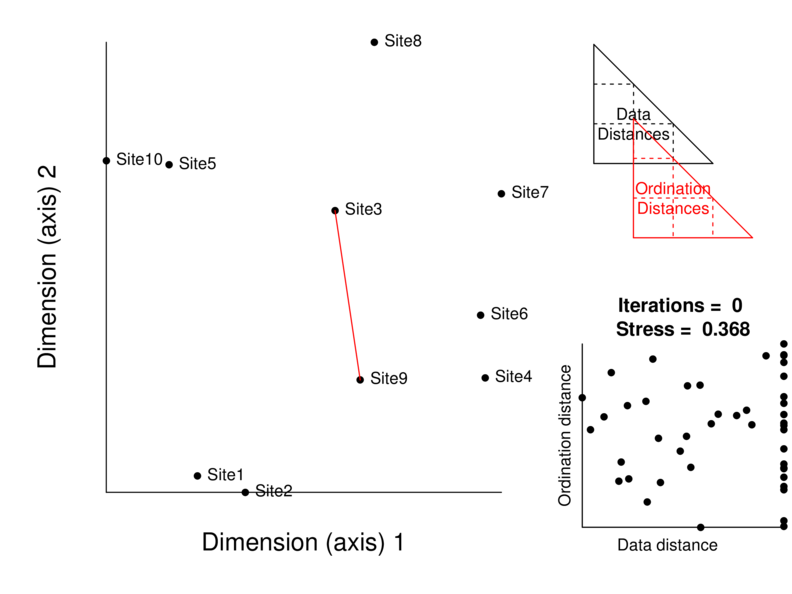
Multidimensional scaling
measure Kruskal’s stress
Multidimensional scaling
iterate - gradient descent
Multidimensional scaling
continual to iterate
Multidimensional scaling
continual (stopping criteria)
Multidimensional scaling
Stopping criteria
convergence tolerance (stress below a threshold)
the stress ratio (improvement in stress)
maximum iterations
Ideally stress < 0.2
Multidimensional scaling
Final configuration
axes have no real meaning
operate together to create ordination space
orientation of points is arbitrary
Multidimensional scaling
Starting contiguration
completely random
based on eigen-analysis
repeated random starts
Multidimensional scaling
Procrustes rotation
Multidimensional scaling
Procrustes rotation
Root mean square error (rmse)
rmse < 0.01, and no one > 0.005
stopping criteria
Multidimensional scaling
metaMDS()
transform and scale
generates dissimilarity
PCoA for starting config
up to 20 random starts
procrustes used to determine final config
final scores scaled
Multidimensional scaling
> library (vegan)
> data.nmds <- metaMDS (data[,-1 ])Square root transformation
Wisconsin double standardization
Run 0 stress 9.575935e-05
Run 1 stress 9.419141e-05
... New best solution
... procrustes: rmse 0.02551179 max resid 0.04216078
Run 2 stress 0.0005652599
... procrustes: rmse 0.1885613 max resid 0.2962192
Run 3 stress 0.0003846328
... procrustes: rmse 0.128145 max resid 0.2569595
Run 4 stress 0.1071568
Run 5 stress 0.08642
Run 6 stress 0.0002599177
... procrustes: rmse 0.1766213 max resid 0.2933088
Run 7 stress 9.39752e-05
... New best solution
... procrustes: rmse 0.1050724 max resid 0.2432038
Run 8 stress 9.692363e-05
... procrustes: rmse 0.101578 max resid 0.2286374
Run 9 stress 9.636878e-05
... procrustes: rmse 0.03799946 max resid 0.06399347
Run 10 stress 9.756922e-05
... procrustes: rmse 0.09787159 max resid 0.2172152
Run 11 stress 9.903768e-05
... procrustes: rmse 0.1377402 max resid 0.2070494
Run 12 stress 0.07877608
Run 13 stress 9.67808e-05
... procrustes: rmse 0.105466 max resid 0.2688797
Run 14 stress 0.00122647
Run 15 stress 0.002519856
Run 16 stress 9.401245e-05
... procrustes: rmse 0.01935187 max resid 0.04030324
Run 17 stress 9.207175e-05
... New best solution
... procrustes: rmse 0.1056994 max resid 0.2401655
Run 18 stress 0.00139044
Run 19 stress 9.781088e-05
... procrustes: rmse 0.1089896 max resid 0.25471
Run 20 stress 9.529673e-05
... procrustes: rmse 0.09327002 max resid 0.1897497
Multidimensional scaling
> data.nmds
Call:
metaMDS(comm = data[, -1])
global Multidimensional Scaling using monoMDS
Data: wisconsin(sqrt(data[, -1]))
Distance: bray
Dimensions: 2
Stress: 9.207175e-05
Stress type 1, weak ties
No convergent solutions - best solution after 20 tries
Scaling: centring, PC rotation, halfchange scaling
Species: expanded scores based on 'wisconsin(sqrt(data[, -1]))'
Multidimensional scaling
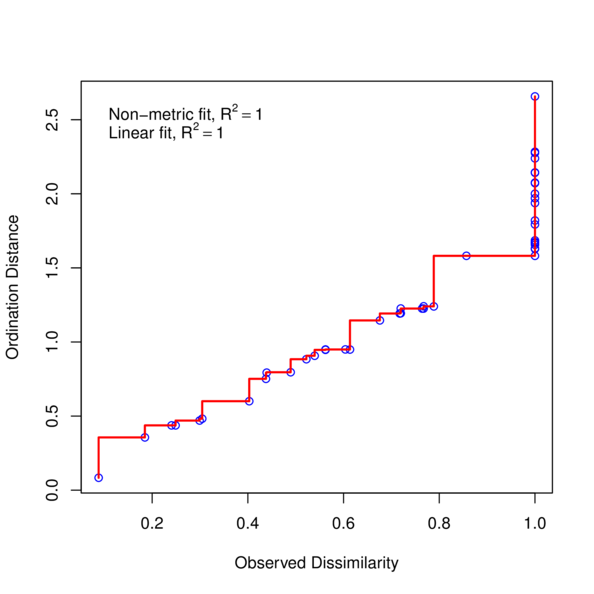
Shepard diagram
> stressplot (data.nmds)
Multidimensional scaling
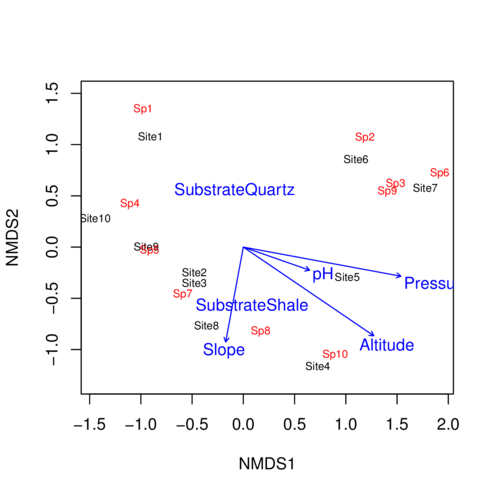
Ordination plot
> ordiplot (data.nmds, type= "text" )
Multidimensional scaling
Environmental fit
rotate axes to planes of maximum variability
observations are not independent
permutation tests - e.g based on \(R^2\)
Multidimensional scaling
Environmental fit
> ordiplot (data.nmds, type= "text" )
> data.envfit <- envfit (data.nmds,enviro[,-1 ])
> plot (data.envfit)
Multidimensional scaling
Environmental fit
> data.envfit
***VECTORS
NMDS1 NMDS2 r2 Pr(>r)
pH 0.94416 -0.32948 0.1605 0.551
Slope -0.18315 -0.98308 0.3033 0.264
Pressure 0.98333 -0.18182 0.8361 0.005 **
Altitude 0.82690 -0.56235 0.8107 0.002 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Permutation: free
Number of permutations: 999
***FACTORS:
Centroids:
NMDS1 NMDS2
SubstrateQuartz -0.0849 0.5622
SubstrateShale 0.0849 -0.5622
Goodness of fit:
r2 Pr(>r)
Substrate 0.2178 0.144
Permutation: free
Number of permutations: 999
Environmental correlates
Sites
Sp1
Sp2
Sp3
Sp4
Sp5
Sp6
Sp7
Sp8
Sp9
Sp10
Site1
Site1
5
0
0
65
5
0
0
0
0
0
Site2
Site2
0
0
0
25
39
0
6
23
0
0
Site3
Site3
0
0
0
6
42
0
6
31
0
0
Site4
Site4
0
0
0
0
0
0
0
40
0
14
Site5
Site5
0
0
6
0
0
0
0
34
18
12
Site6
Site6
0
29
12
0
0
0
0
0
22
0
Site7
Site7
0
0
21
0
0
5
0
0
20
0
Site8
Site8
0
0
0
0
13
0
6
37
0
0
Site9
Site9
0
0
0
60
47
0
4
0
0
0
Site10
Site10
0
0
0
72
34
0
0
0
0
0
Environmental correlates
Site
pH
Slope
Pressure
Altitude
Substrate
Site1
6
4
101325
2
Quartz
Site2
7
9
101352
510
Shale
Site3
7
9
101356
546
Shale
Site4
7
7
101372
758
Shale
Site5
7
6
101384
813
Shale
Site6
8
8
101395
856
Quartz
Site7
8
0
101396
854
Quartz
Site8
7
12
101370
734
Shale
Site9
8
8
101347
360
Quartz
Site10
6
2
101345
356
Quartz
Environmental correlates
PCA/CA
relate envir to first few axes
MDS with envfit
relate envir to reduced optimized axes
All just relate envir to SOME patterns of community data
Mantel test
correlation of two distance matrices
Mantel test
> library (vegan)
> data.dist <- vegdist (wisconsin (sqrt (data[,-1 ])),"bray" )
> data.dist Site1 Site2 Site3 Site4 Site5 Site6
Site2 0.67587556
Site3 0.76402116 0.08814559
Site4 1.00000000 0.76728722 0.71732569
Site5 1.00000000 0.76728722 0.71950051 0.43782491
Site6 1.00000000 1.00000000 1.00000000 1.00000000 0.56217509
Site7 1.00000000 1.00000000 1.00000000 1.00000000 0.56217509 0.40287938
Site8 0.85671371 0.24898448 0.18481903 0.61338910 0.71950051 1.00000000
Site9 0.52225127 0.24045299 0.30461844 1.00000000 1.00000000 1.00000000
Site10 0.43930743 0.53960530 0.60377075 1.00000000 1.00000000 1.00000000
Site7 Site8 Site9
Site2
Site3
Site4
Site5
Site6
Site7
Site8 1.00000000
Site9 1.00000000 0.48943747
Site10 1.00000000 0.78858978 0.29915230
Mantel test
Environmental distances
> library (vegan)
> enviro1 <- within (enviro,
+ Substrate<-as.numeric (Substrate))
> env.dist <- vegdist (decostand (enviro1[,-1 ],
+ "standardize" ),"euclid" )
> env.dist 1 2 3 4 5 6 7
2 3.3229931
3 3.4352275 0.3112630
4 4.2003679 1.4095427 1.1199511
5 4.6243094 2.1315293 1.8280978 0.7722349
6 4.9177360 3.1669299 2.9561936 2.3097932 2.1404251
7 4.9080429 4.0132037 3.7487386 3.0169364 2.5115880 2.2216568
8 4.5387150 1.4066733 1.3097554 1.2431118 1.8378136 2.5328911 3.9603544
9 3.9400812 3.2272044 3.1427473 3.3444512 3.5238311 3.0372913 3.7500941
10 1.7177533 3.0391765 3.0117638 3.3462020 3.5745559 3.9177255 3.4387521
8 9
2
3
4
5
6
7
8
9 3.4268548
10 4.0495542 3.7782159
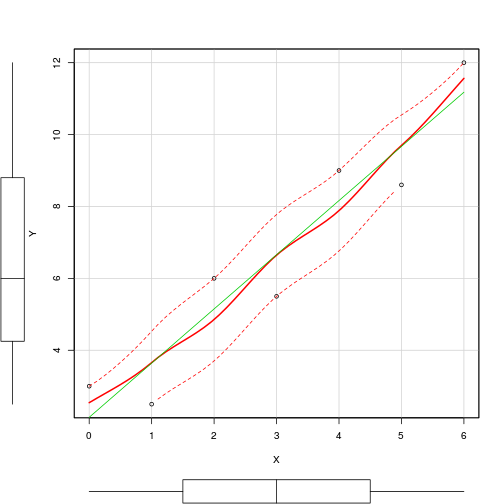
Mantel test
> plot (data.dist, env.dist)
Mantel test
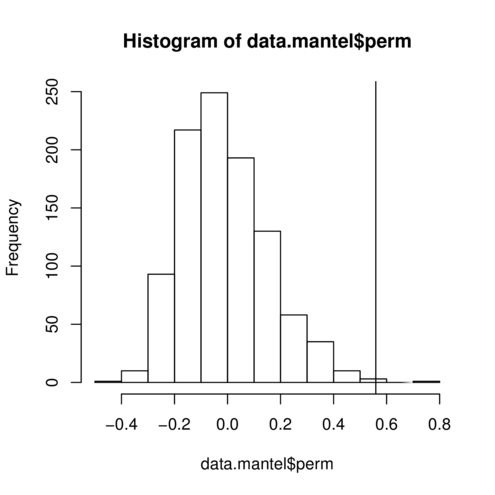
> data.mantel <- mantel (data.dist, env.dist, perm= 1000 )
> data.mantel
Mantel statistic based on Pearson's product-moment correlation
Call:
mantel(xdis = data.dist, ydis = env.dist, permutations = 1000)
Mantel statistic r: 0.5593
Significance: 0.003996
Upper quantiles of permutations (null model):
90% 95% 97.5% 99%
0.209 0.297 0.357 0.428
Permutation: free
Number of permutations: 1000
Mantel test
> hist (data.mantel$perm)
> abline (v= data.mantel$statistic)
Best environmental subsets
looks at combinations of environmental predictors
analogous to model selection
not an inference test
Best environmental subsets
> # convert the Substrate factor to a numeric via dummy coding
> # function definition at the start of this Tutorial
> dummy <- function(x) {
+ nms <- colnames (x)
+ ff <- eval (parse (text= paste ("~" ,paste (nms,collapse= "+" ))))
+ mm <- model.matrix (ff,x)
+ nms <- colnames (mm)
+ mm <- as.matrix (mm[,-1 ])
+ colnames (mm) <- nms[-1 ]
+ mm
+ }
> enviro1 <- dummy (enviro[,-1 ])
> enviro1 pH Slope Pressure Altitude SubstrateShale
1 6.1 4.2 101325 2 0
2 6.7 9.2 101352 510 1
3 6.8 8.6 101356 546 1
4 7.0 7.4 101372 758 1
5 7.2 5.8 101384 813 1
6 7.5 8.4 101395 856 0
7 7.5 0.5 101396 854 0
8 7.0 11.8 101370 734 1
9 8.4 8.2 101347 360 0
10 6.2 1.5 101345 356 0
Best environmental subsets
> library (car)
> vif (lm (1 :nrow (enviro1) ~ pH+Slope+Pressure+Altitude+SubstrateShale, data= data.frame (enviro1))) pH Slope Pressure
1.976424 2.187796 45.804821
Altitude SubstrateShale
52.754368 5.118418 > vif (lm (1 :nrow (enviro1) ~ pH+Slope+Altitude+SubstrateShale, data= data.frame (enviro1))) pH Slope Altitude
1.968157 2.116732 1.830014
SubstrateShale
2.636302
Best environmental subsets
> # Pressure is in column three
> data.bioenv <- bioenv (wisconsin (sqrt (data[,-1 ])), decostand (enviro1[,-3 ],"standardize" ))
> data.bioenv
Call:
bioenv(comm = wisconsin(sqrt(data[, -1])), env = decostand(enviro1[, -3], "standardize"))
Subset of environmental variables with best correlation to community data.
Correlations: spearman
Dissimilarities: bray
Metric: euclidean
Best model has 1 parameters (max. 4 allowed):
Altitude
with correlation 0.5514973
Permutation Multivariate Analysis of Variance
adonis
partitions the total sums of squares
contribution of each predictor
permutation test of inference
Permutation Multivariate Analysis of Variance
adonis
> data.adonis <- adonis (data.dist ~ pH + Slope + Altitude +
+ Substrate, data= enviro)
> data.adonis
Call:
adonis(formula = data.dist ~ pH + Slope + Altitude + Substrate, data = enviro)
Permutation: free
Number of permutations: 999
Terms added sequentially (first to last)
Df SumsOfSqs MeanSqs F.Model R2 Pr(>F)
pH 1 0.21899 0.21899 1.6290 0.07768 0.209
Slope 1 0.55960 0.55960 4.1628 0.19850 0.015 *
Altitude 1 0.98433 0.98433 7.3223 0.34916 0.002 **
Substrate 1 0.38404 0.38404 2.8568 0.13623 0.057 .
Residuals 5 0.67215 0.13443 0.23842
Total 9 2.81911 1.00000
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Worked Examples
> data <- read.csv ('../data/data.csv' , strip.white= TRUE )
> head (data) Sites Sp1 Sp2 Sp3 Sp4 Sp5 Sp6 Sp7 Sp8 Sp9 Sp10
1 Site1 5 0 0 65 5 0 0 0 0 0
2 Site2 0 0 0 25 39 0 6 23 0 0
3 Site3 0 0 0 6 42 0 6 31 0 0
4 Site4 0 0 0 0 0 0 0 40 0 14
5 Site5 0 0 6 0 0 0 0 34 18 12
6 Site6 0 29 12 0 0 0 0 0 22 0> enviro <- read.csv ('../data/enviro.csv' , strip.white= TRUE )
> head (enviro) Site pH Slope Pressure Altitude Substrate
1 Site1 6.1 4.2 101325 2 Quartz
2 Site2 6.7 9.2 101352 510 Shale
3 Site3 6.8 8.6 101356 546 Shale
4 Site4 7.0 7.4 101372 758 Shale
5 Site5 7.2 5.8 101384 813 Shale
6 Site6 7.5 8.4 101395 856 Quartz> data.bc <- vegdist (data[,-1 ], 'bray' )
>
> data.cpscale<-capscale (data.bc~ pH+Slope+Altitude+Substrate, data= enviro)
> plot (data.cpscale)
plot of chunk qmodeExamples
> str (data.cpscale)List of 13
$ call : language capscale(formula = data.bc ~ pH + Slope + Altitude + Substrate, data = enviro)
$ grand.total: logi NA
$ rowsum : logi NA
$ colsum : logi NA
$ tot.chi : num 2.76
$ pCCA : NULL
$ CCA :List of 12
..$ eig : Named num [1:4] 1.266 0.978 0.073 0.03
.. ..- attr(*, "names")= chr [1:4] "CAP1" "CAP2" "CAP3" "CAP4"
..$ u : num [1:10, 1:4] -0.66766 -0.04799 -0.00444 0.24882 0.31907 ...
.. ..- attr(*, "dimnames")=List of 2
.. .. ..$ : chr [1:10] "1" "2" "3" "4" ...
.. .. ..$ : chr [1:4] "CAP1" "CAP2" "CAP3" "CAP4"
..$ v : num [1:6, 1:4] NA NA NA NA NA NA NA NA NA NA ...
.. ..- attr(*, "dimnames")=List of 2
.. .. ..$ : chr [1:6] "Dim1" "Dim2" "Dim3" "Dim4" ...
.. .. ..$ : chr [1:4] "CAP1" "CAP2" "CAP3" "CAP4"
..$ wa : num [1:10, 1:4] -0.4575 -0.1783 -0.0215 0.3049 0.4377 ...
.. ..- attr(*, "dimnames")=List of 2
.. .. ..$ : chr [1:10] "1" "2" "3" "4" ...
.. .. ..$ : chr [1:4] "CAP1" "CAP2" "CAP3" "CAP4"
..$ biplot : num [1:4, 1:4] 0.312 0.138 0.987 0.442 0.256 ...
.. ..- attr(*, "dimnames")=List of 2
.. .. ..$ : chr [1:4] "pH" "Slope" "Altitude" "SubstrateShale"
.. .. ..$ : chr [1:4] "CAP1" "CAP2" "CAP3" "CAP4"
..$ rank : int 4
..$ qrank : int 4
..$ tot.chi : num 2.35
..$ QR :List of 4
.. ..$ qr : num [1:10, 1:4] 2.0159 0.1687 0.1191 0.0198 -0.0794 ...
.. .. ..- attr(*, "dimnames")=List of 2
.. .. .. ..$ : chr [1:10] "1" "2" "3" "4" ...
.. .. .. ..$ : chr [1:4] "pH" "Slope" "Altitude" "SubstrateShale"
.. ..$ rank : int 4
.. ..$ qraux: num [1:4] 1.47 1.31 1.04 1.19
.. ..$ pivot: int [1:4] 1 2 3 4
.. ..- attr(*, "class")= chr "qr"
..$ envcentre: Named num [1:4] 7.04 6.56 578.9 0.5
.. ..- attr(*, "names")= chr [1:4] "pH" "Slope" "Altitude" "SubstrateShale"
..$ Xbar : num [1:10, 1:6] -1.261 -0.808 -0.401 0.787 1.424 ...
.. ..- attr(*, "dimnames")=List of 2
.. .. ..$ : chr [1:10] "1" "2" "3" "4" ...
.. .. ..$ : chr [1:6] "Dim1" "Dim2" "Dim3" "Dim4" ...
.. ..- attr(*, "scaled:center")= Named num [1:6] 1.80e-16 1.11e-16 -2.76e-16 7.79e-17 1.06e-15 ...
.. .. ..- attr(*, "names")= chr [1:6] "Dim1" "Dim2" "Dim3" "Dim4" ...
..$ centroids: num [1:2, 1:4] -0.14 0.14 0.283 -0.283 0.015 ...
.. ..- attr(*, "dimnames")=List of 2
.. .. ..$ : chr [1:2] "SubstrateQuartz" "SubstrateShale"
.. .. ..$ : chr [1:4] "CAP1" "CAP2" "CAP3" "CAP4"
$ CA :List of 9
..$ eig : Named num [1:5] 0.35573 0.1688 0.04051 0.02265 0.00193
.. ..- attr(*, "names")= chr [1:5] "MDS1" "MDS2" "MDS3" "MDS4" ...
..$ u : num [1:10, 1:5] -0.567 0.262 0.28 -0.269 -0.342 ...
.. ..- attr(*, "dimnames")=List of 2
.. .. ..$ : chr [1:10] "1" "2" "3" "4" ...
.. .. ..$ : chr [1:5] "MDS1" "MDS2" "MDS3" "MDS4" ...
..$ v : num [1:6, 1:5] NA NA NA NA NA NA NA NA NA NA ...
.. ..- attr(*, "dimnames")=List of 2
.. .. ..$ : chr [1:6] "Dim1" "Dim2" "Dim3" "Dim4" ...
.. .. ..$ : chr [1:5] "MDS1" "MDS2" "MDS3" "MDS4" ...
..$ rank : int 5
..$ tot.chi : num 0.59
..$ Xbar : num [1:10, 1:6] 0.853 -0.373 -0.123 0.145 0.528 ...
.. ..- attr(*, "dimnames")=List of 2
.. .. ..$ : chr [1:10] "1" "2" "3" "4" ...
.. .. ..$ : chr [1:6] "Dim1" "Dim2" "Dim3" "Dim4" ...
.. ..- attr(*, "scaled:center")= Named num [1:6] 1.80e-16 1.11e-16 -2.76e-16 7.79e-17 1.06e-15 ...
.. .. ..- attr(*, "names")= chr [1:6] "Dim1" "Dim2" "Dim3" "Dim4" ...
..$ imaginary.chi : num -0.176
..$ imaginary.rank : int 3
..$ imaginary.u.eig: num [1:10, 1:3] -0.00467 -0.04869 0.01533 -0.00407 -0.01243 ...
.. ..- attr(*, "dimnames")=List of 2
.. .. ..$ : chr [1:10] "1" "2" "3" "4" ...
.. .. ..$ : NULL
$ method : chr "capscale"
$ inertia : chr "squared Bray distance"
$ terms :Classes 'terms', 'formula' length 3 data.bc ~ pH + Slope + Altitude + Substrate
.. ..- attr(*, "variables")= language list(data.bc, pH, Slope, Altitude, Substrate)
.. ..- attr(*, "factors")= int [1:5, 1:4] 0 1 0 0 0 0 0 1 0 0 ...
.. .. ..- attr(*, "dimnames")=List of 2
.. .. .. ..$ : chr [1:5] "data.bc" "pH" "Slope" "Altitude" ...
.. .. .. ..$ : chr [1:4] "pH" "Slope" "Altitude" "Substrate"
.. ..- attr(*, "term.labels")= chr [1:4] "pH" "Slope" "Altitude" "Substrate"
.. ..- attr(*, "specials")=Dotted pair list of 1
.. .. ..$ Condition: NULL
.. ..- attr(*, "order")= int [1:4] 1 1 1 1
.. ..- attr(*, "intercept")= int 1
.. ..- attr(*, "response")= int 1
.. ..- attr(*, ".Environment")=<environment: R_GlobalEnv>
$ terminfo :List of 3
..$ terms :Classes 'terms', 'formula' length 2 ~pH + Slope + Altitude + Substrate
.. .. ..- attr(*, "variables")= language list(pH, Slope, Altitude, Substrate)
.. .. ..- attr(*, "factors")= int [1:4, 1:4] 1 0 0 0 0 1 0 0 0 0 ...
.. .. .. ..- attr(*, "dimnames")=List of 2
.. .. .. .. ..$ : chr [1:4] "pH" "Slope" "Altitude" "Substrate"
.. .. .. .. ..$ : chr [1:4] "pH" "Slope" "Altitude" "Substrate"
.. .. ..- attr(*, "term.labels")= chr [1:4] "pH" "Slope" "Altitude" "Substrate"
.. .. ..- attr(*, "order")= int [1:4] 1 1 1 1
.. .. ..- attr(*, "intercept")= int 1
.. .. ..- attr(*, "response")= num 0
.. .. ..- attr(*, ".Environment")=<environment: R_GlobalEnv>
..$ xlev :List of 1
.. ..$ Substrate: chr [1:2] "Quartz" "Shale"
..$ ordered: Named logi [1:4] FALSE FALSE FALSE FALSE
.. ..- attr(*, "names")= chr [1:4] "pH" "Slope" "Altitude" "Substrate"
$ adjust : num 3
- attr(*, "class")= chr [1:3] "capscale" "rda" "cca"> data.cmdscale <- cmdscale (data.bc, k= 2 )
> plot (data.cmdscale)
plot of chunk qmodeExamples
> data.scores <- scores (data.cmdscale)