Multiple Linear Regression
Additive model
\(growth = intercept + temperature + nitrogen\)
\(y_i=\beta_0+\beta_1x_{i1}+\beta_2x_{i2}+...+\beta_jx_{ij}+\epsilon_i\)
| Y | X1 | X2 |
|---|---|---|
| 3 | 22.7 | 0.9 |
| 2.5 | 23.7 | 0.5 |
| 6 | 25.7 | 0.6 |
| 5.5 | 29.1 | 0.7 |
| 9 | 22 | 0.8 |
| 8.6 | 29 | 1.3 |
| 12 | 29.4 | 1 |
Multiple Linear Regression
Additive model
\[ \begin{alignat*}{1} 3~&=~\beta_0 ~&+~(\beta_1\times22.7) ~&+~(\beta_2\times0.9)~&+~\varepsilon_1\\ 2.5~&=~\beta_0 ~&+~(\beta_1\times23.7) ~&+~(\beta_2\times0.5)~&+~\varepsilon_2\\ 6~&=~\beta_0 ~&+~(\beta_1\times25.7) ~&+~(\beta_2\times0.6)~&+~\varepsilon_3\\ 5.5~&=~\beta_0 ~&+~(\beta_1\times29.1) ~&+~(\beta_2\times0.7)~&+~\varepsilon_4\\ 9~&=~\beta_0 ~&+~(\beta_1\times22) ~&+~(\beta_2\times0.8)~&+~\varepsilon_5\\ 8.6~&=~\beta_0 ~&+~(\beta_1\times29) ~&+~(\beta_2\times1.3)~&+~\varepsilon_6\\ 12~&=~\beta_0 ~&+~(\beta_1\times29.4) ~&+~(\beta_2\times1)~&+~\varepsilon_7\\ \end{alignat*} \]
Multiple Linear Regression
Multiplicative model
\[growth = intercept + temp + nitro + temp\times nitro\]
\[y_i=\beta_0+\beta_1x_{i1}+\beta_2x_{i2}+\beta_3x_{i1}x_{i2}+...+\epsilon_i\]
Multiple Linear Regression
Multiplicative model
\[ \begin{alignat*}{1} 3~&=~\beta_0 ~&+~(\beta_1\times22.7) ~&+~(\beta_2\times0.9)~&+~(\beta_3\times22.7\times0.9)~&+~\varepsilon_1\\ 2.5~&=~\beta_0 ~&+~(\beta_1\times23.7) ~&+~(\beta_2\times0.5)~&+~(\beta_3\times23.7\times0.5)~&+~\varepsilon_2\\ 6~&=~\beta_0 ~&+~(\beta_1\times25.7) ~&+~(\beta_2\times0.6)~&+~(\beta_3\times25.7\times0.6)~&+~\varepsilon_3\\ 5.5~&=~\beta_0 ~&+~(\beta_1\times29.1) ~&+~(\beta_2\times0.7)~&+~(\beta_3\times29.1\times0.7)~&+~\varepsilon_4\\ 9~&=~\beta_0 ~&+~(\beta_1\times22) ~&+~(\beta_2\times0.8)~&+~(\beta_3\times22\times0.8)~&+~\varepsilon_5\\ 8.6~&=~\beta_0 ~&+~(\beta_1\times29) ~&+~(\beta_2\times1.3)~&+~(\beta_3\times29\times1.3)~&+~\varepsilon_6\\ 12~&=~\beta_0 ~&+~(\beta_1\times29.4) ~&+~(\beta_2\times1)~&+~(\beta_3\times29.4\times1)~&+~\varepsilon_7\\ \end{alignat*} \]
Centering data
Multiple Linear Regression
Centering
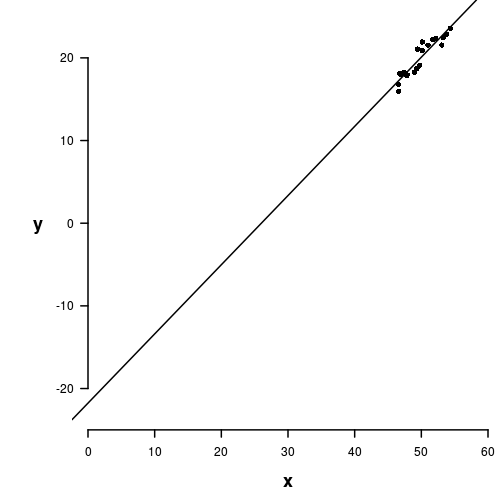
Multiple Linear Regression
Centering
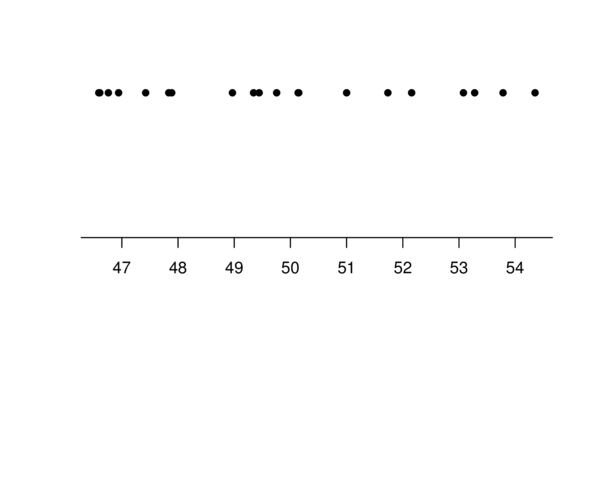
Multiple Linear Regression
Centering
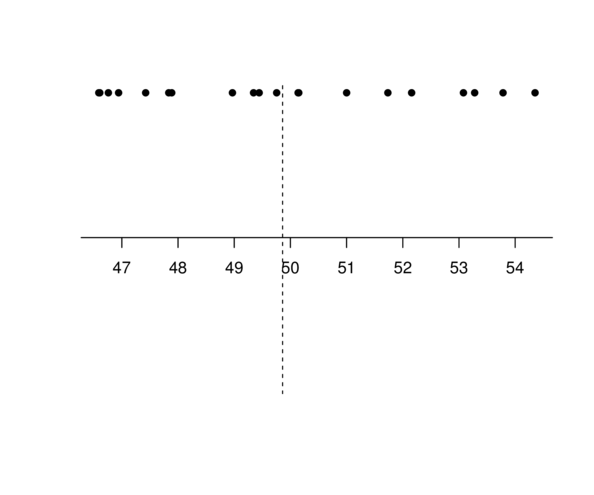
Multiple Linear Regression
Centering
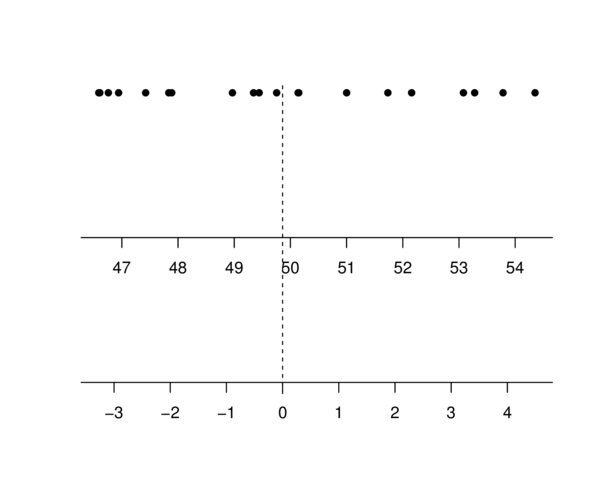
Multiple Linear Regression
Centering
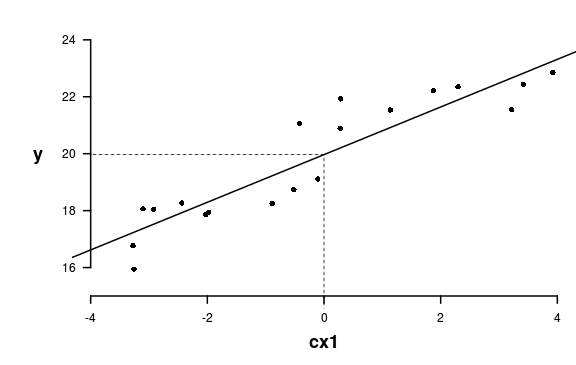
Assumptions
Multiple Linear Regression
Assumptions
Normality, homog., linearity
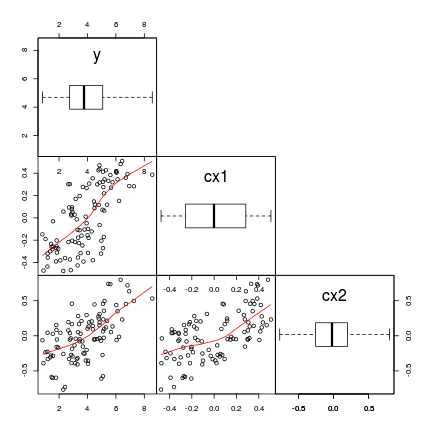
Multiple Linear Regression
Assumptions
(multi)collinearity

Multiple Linear Regression
Variance inflation
Strength of a relationship \[ R^2 \] Strong when \(R^2 \ge 0.8\)
Multiple Linear Regression
Variance inflation
\[ var.inf = \frac{1}{1-R^2} \] Collinear when \(var.inf >= 5\)
Some prefer \(>3\)
Multiple Linear Regression
Assumptions
(multi)collinearity
library(car)
# additive model - scaled predictors
vif(lm(y ~ cx1 + cx2, data)) cx1 cx2
1.743817 1.743817 Multiple Linear Regression
Assumptions
(multi)collinearity
library(car)
# additive model - scaled predictors
vif(lm(y ~ cx1 + cx2, data)) cx1 cx2
1.743817 1.743817 # multiplicative model - raw predictors
vif(lm(y ~ x1 * x2, data)) x1 x2 x1:x2
7.259729 5.913254 16.949468 Multiple Linear Regression
Assumptions
# multiplicative model - raw predictors
vif(lm(y ~ x1 * x2, data)) x1 x2 x1:x2
7.259729 5.913254 16.949468 # multiplicative model - scaled predictors
vif(lm(y ~ cx1 * cx2, data)) cx1 cx2 cx1:cx2
1.769411 1.771994 1.018694 Multiple linear models in R
Model fitting
Additive model
\(y_i=\beta_0+\beta_1x_{i1}+\beta_2x_{i2}+\epsilon_i\)
data.add.lm <- lm(y~cx1+cx2, data)Model fitting
Additive model
\(y_i=\beta_0+\beta_1x_{i1}+\beta_2x_{i2}+\epsilon_i\)
data.add.lm <- lm(y~cx1+cx2, data)
Multiplicative model
\(y_i=\beta_0+\beta_1x_{i1}+\beta_2x_{i2}+\beta_3x_{i1}x_{i2}+\epsilon_i\)
data.mult.lm <- lm(y~cx1+cx2+cx1:cx2, data)
#OR
data.mult.lm <- lm(y~cx1*cx2, data)Model evaluation
Additive model
plot(data.add.lm)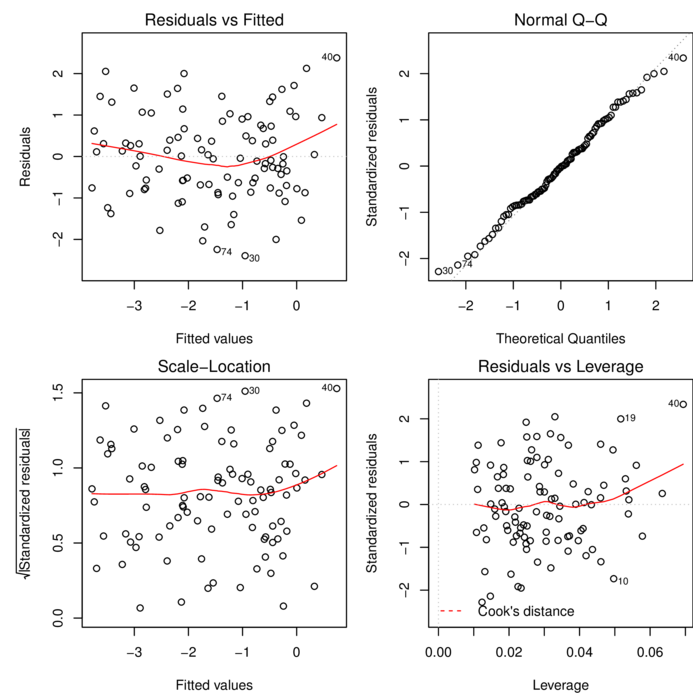
Model evaluation
Multiplicative model
plot(data.mult.lm)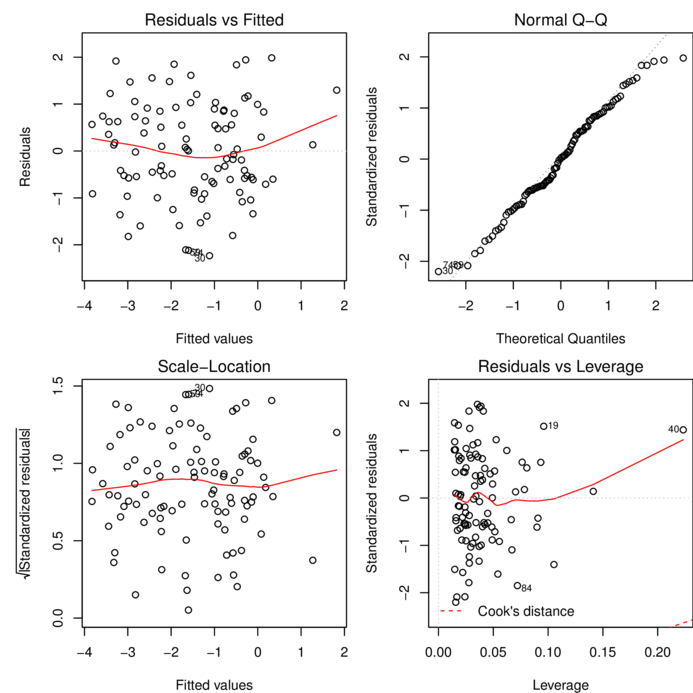
Model summary
Additive model
summary(data.add.lm)
Call:
lm(formula = y ~ cx1 + cx2, data = data)
Residuals:
Min 1Q Median 3Q Max
-2.39418 -0.75888 -0.02463 0.73688 2.37938
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -1.5161 0.1055 -14.364 < 2e-16 ***
cx1 2.5749 0.4683 5.499 3.1e-07 ***
cx2 -4.0475 0.3734 -10.839 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.055 on 97 degrees of freedom
Multiple R-squared: 0.5567, Adjusted R-squared: 0.5476
F-statistic: 60.91 on 2 and 97 DF, p-value: < 2.2e-16Model summary
Additive model
confint(data.add.lm) 2.5 % 97.5 %
(Intercept) -1.725529 -1.306576
cx1 1.645477 3.504300
cx2 -4.788628 -3.306308Model summary
Multiplicative model
summary(data.mult.lm)
Call:
lm(formula = y ~ cx1 * cx2, data = data)
Residuals:
Min 1Q Median 3Q Max
-2.23364 -0.62188 0.01763 0.80912 1.98568
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -1.6995 0.1228 -13.836 < 2e-16 ***
cx1 2.7232 0.4571 5.957 4.22e-08 ***
cx2 -4.1716 0.3648 -11.435 < 2e-16 ***
cx1:cx2 2.5283 0.9373 2.697 0.00826 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.023 on 96 degrees of freedom
Multiple R-squared: 0.588, Adjusted R-squared: 0.5751
F-statistic: 45.66 on 3 and 96 DF, p-value: < 2.2e-16Graphical summaries
Additive model
library(effects)
plot(effect("cx1",data.add.lm, partial.residuals=TRUE))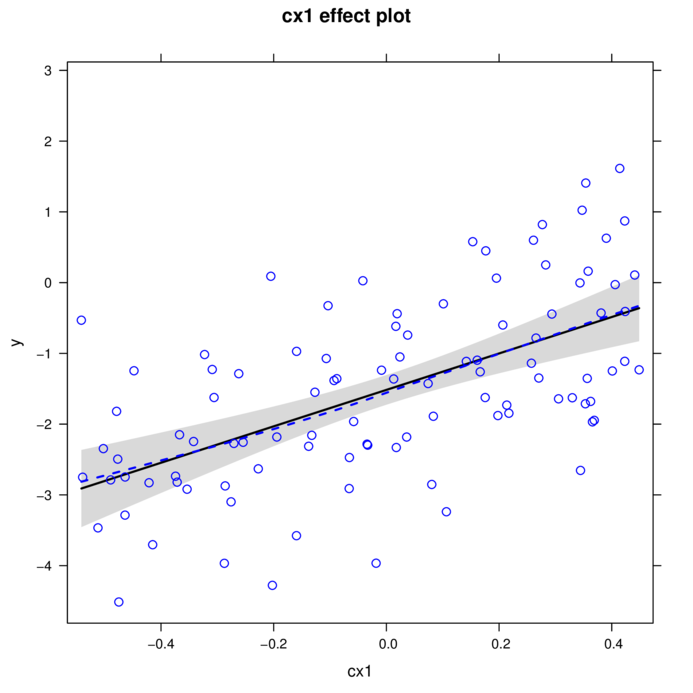
Graphical summaries
Additive model
library(effects)
library(ggplot2)
e <- effect("cx1",data.add.lm, xlevels=list(
cx1=seq(-0.4,0.4, len=10)), partial.residuals=TRUE)
newdata <- data.frame(fit=e$fit, cx1=e$x, lower=e$lower,
upper=e$upper)
resids <- data.frame(resid=e$partial.residuals.raw,
cx1=e$data$cx1) Error in data.frame(resid = e$partial.residuals.raw, cx1 = e$data$cx1): arguments imply differing number of rows: 0, 100ggplot(newdata, aes(y=fit, x=cx1)) +
geom_point(data=resids, aes(y=resid, x=cx1))+
geom_ribbon(aes(ymin=lower, ymax=upper), fill='blue',
alpha=0.2)+
geom_line()+theme_classic()Error in fortify(data): object 'resids' not foundGraphical summaries
Additive model
library(effects)
plot(allEffects(data.add.lm))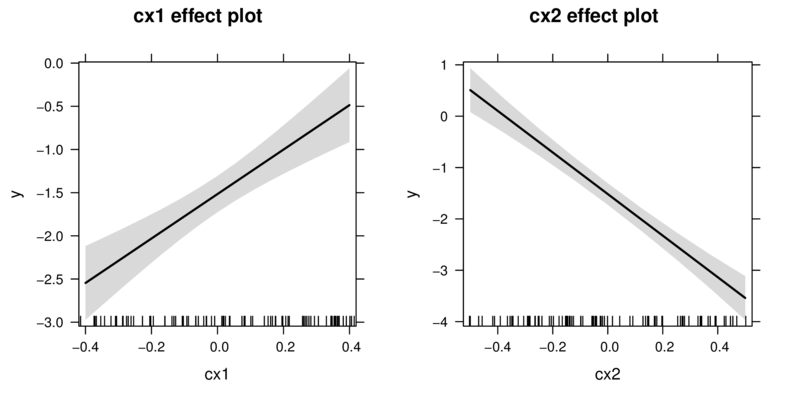
Graphical summaries
Multiplicative model
library(effects)
plot(allEffects(data.mult.lm))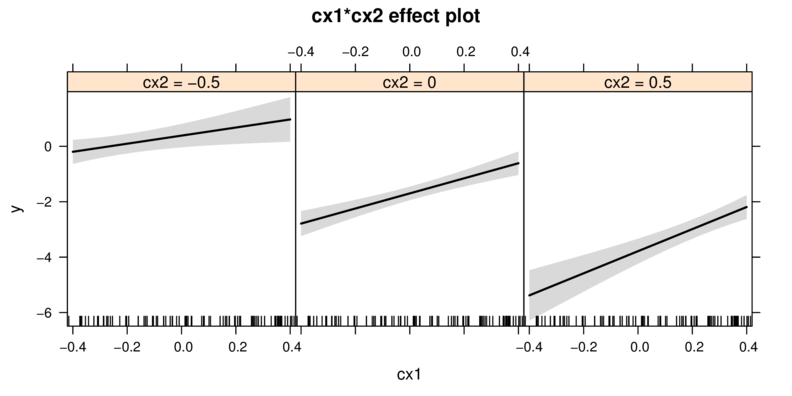
Graphical summaries
Multiplicative model
library(effects)
plot(Effect(focal.predictors=c("cx1","cx2"),data.mult.lm))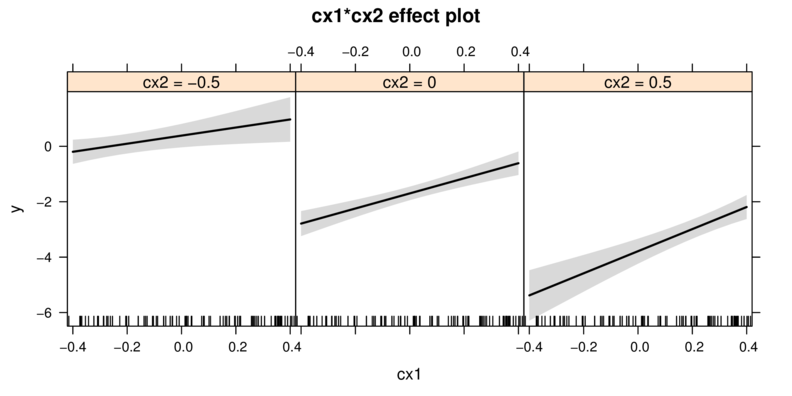
Model selection
How good is a model?
“All models are wrong, but some are useful” George E. P. Box
Criteria
- \(R^2\) - no
- Information criteria
- AIC, AICc
- penalize for complexity
Model selection
Candidates
AIC(data.add.lm, data.mult.lm) df AIC
data.add.lm 4 299.5340
data.mult.lm 5 294.2283library(MuMIn)
AICc(data.add.lm, data.mult.lm) df AICc
data.add.lm 4 299.9551
data.mult.lm 5 294.8666Model selection
Dredging
library(MuMIn)
data.mult.lm <- lm(y~cx1*cx2, data, na.action=na.fail)
dredge(data.mult.lm, rank="AICc", trace=TRUE)0 : lm(formula = y ~ 1, data = data, na.action = na.fail)
1 : lm(formula = y ~ cx1 + 1, data = data, na.action = na.fail)
2 : lm(formula = y ~ cx2 + 1, data = data, na.action = na.fail)
3 : lm(formula = y ~ cx1 + cx2 + 1, data = data, na.action = na.fail)
7 : lm(formula = y ~ cx1 + cx2 + cx1:cx2 + 1, data = data, na.action = na.fail)Global model call: lm(formula = y ~ cx1 * cx2, data = data, na.action = na.fail)
---
Model selection table
(Int) cx1 cx2 cx1:cx2 df logLik AICc delta weight
8 -1.699 2.7230 -4.172 2.528 5 -142.114 294.9 0.00 0.927
4 -1.516 2.5750 -4.047 4 -145.767 300.0 5.09 0.073
3 -1.516 -2.706 3 -159.333 324.9 30.05 0.000
1 -1.516 2 -186.446 377.0 82.15 0.000
2 -1.516 -0.7399 3 -185.441 377.1 82.27 0.000
Models ranked by AICc(x) Multiple Linear Regression
Model averaging
library(MuMIn)
data.dredge<-dredge(data.mult.lm, rank="AICc")
model.avg(data.dredge, subset=delta<20)
Call:
model.avg(object = data.dredge, subset = delta < 20)
Component models:
'123' '12'
Coefficients:
(Intercept) cx1 cx2 cx1:cx2
full -1.686125 2.712397 -4.162525 2.344227
subset -1.686125 2.712397 -4.162525 2.528328Multiple Linear Regression
Model selection
Or more preferable:
- identify 10-15 candidate models
- compare these via AIC (etc)
Worked Examples
Worked examples
| Format of loyn.csv data file | |||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|

|
loyn <- read.csv('../data/loyn.csv', strip.white=T)
head(loyn) ABUND AREA YR.ISOL DIST LDIST GRAZE ALT
1 5.3 0.1 1968 39 39 2 160
2 2.0 0.5 1920 234 234 5 60
3 1.5 0.5 1900 104 311 5 140
4 17.1 1.0 1966 66 66 3 160
5 13.8 1.0 1918 246 246 5 140
6 14.1 1.0 1965 234 285 3 130Worked Examples
Question: what effects do fragmentation variables have on the abundance of forest birds
Linear model: