Workshop 9.15a: Heterogeneity
Murray Logan
27-04-2014
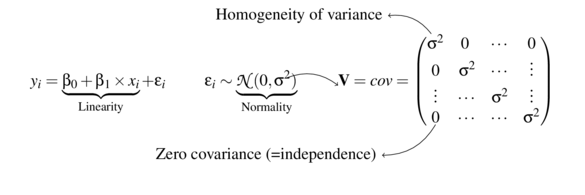
Linear modelling assumptions
\[
\begin{align*}
y_{i} &= \beta_0 + \beta_1 \times x_{i} + \varepsilon_i\\
\epsilon_i&\sim\mathcal{N}(0, \sigma^2) \\
\end{align*}
\]
Linear modelling assumptions
\[
\begin{align*}
y_{i} &= \beta_0 + \beta_1 \times x_{i} + \varepsilon_i\\
\epsilon_i&\sim\mathcal{N}(0, \sigma^2) \\
\end{align*}
\]
Dealing with Heterogeneity
41.9
1
48.5
2
43
3
51.4
4
51.2
5
37.7
6
50.7
7
65.1
8
51.7
9
38.9
10
70.6
11
51.4
12
62.7
13
34.9
14
95.3
15
63.9
16
Dealing with Heterogeneity
data1 <- read.csv ('../data/D1.csv' )
summary (data1) y x
Min. :34.9 Min. : 1.00
1st Qu.:42.7 1st Qu.: 4.75
Median :51.3 Median : 8.50
Mean :53.7 Mean : 8.50
3rd Qu.:63.0 3rd Qu.:12.25
Max. :95.3 Max. :16.00
\[
\begin{align*}
y_{i} &= \beta_0 + \beta_1 \times x_{i} + \varepsilon_i\\
\epsilon_i&\sim\mathcal{N}(0, \sigma^2) \\
\end{align*}
\]
estimate \(\beta_0\), \(\beta_1\) and \(\sigma^2\)
Dealing with Heterogeneity
Dealing with Heterogeneity
Dealing with Heterogeneity
\[
\mathbf{V} = cov =
\underbrace{
\begin{pmatrix}
\sigma^2&0&0&0&0&0&0&0&0&0&0&0&0&0&0\\
0&\sigma^2&0&0&0&0&0&0&0&0&0&0&0&0&0\\
0&0&\sigma^2&0&0&0&0&0&0&0&0&0&0&0&0\\
0&0&0&\sigma^2&0&0&0&0&0&0&0&0&0&0&0\\
0&0&0&0&\sigma^2&0&0&0&0&0&0&0&0&0&0\\
0&0&0&0&0&\sigma^2&0&0&0&0&0&0&0&0&0\\
0&0&0&0&0&0&\sigma^2&0&0&0&0&0&0&0&0\\
0&0&0&0&0&0&0&\sigma^2&0&0&0&0&0&0&0\\
0&0&0&0&0&0&0&0&\sigma^2&0&0&0&0&0&0\\
0&0&0&0&0&0&0&0&0&\sigma^2&0&0&0&0&0\\
0&0&0&0&0&0&0&0&0&0&\sigma^2&0&0&0&0\\
0&0&0&0&0&0&0&0&0&0&0&\sigma^2&0&0&0\\
0&0&0&0&0&0&0&0&0&0&0&0&\sigma^2&0&0\\
0&0&0&0&0&0&0&0&0&0&0&0&0&\sigma^2&0\\
0&0&0&0&0&0&0&0&0&0&0&0&0&0&\sigma^2\\
\end{pmatrix}}_\text{Variance-covariance matrix}
\]
Dealing with Heterogeneity
\[
\mathbf{V} = \sigma^2\times
\underbrace{
\begin{pmatrix}
1&0&\cdots&0\\
0&1&\cdots&\vdots\\
\vdots&\cdots&1&\vdots\\
0&\cdots&\cdots&1\\
\end{pmatrix}}_\text{Identity matrix} =
\underbrace{
\begin{pmatrix}
\sigma^2&0&\cdots&0\\
0&\sigma^2&\cdots&\vdots\\
\vdots&\cdots&\sigma^2&\vdots\\
0&\cdots&\cdots&\sigma^2\\
\end{pmatrix}}_\text{Variance-covariance matrix}
\]
Dealing with Heterogeneity
variance proportional to X
variance inversely proportional to X
Dealing with Heterogeneity
variance inversely proportional to X
\[
\mathbf{V} = \sigma^2\times X\times
\underbrace{
\begin{pmatrix}
1&0&\cdots&0\\
0&1&\cdots&\vdots\\
\vdots&\cdots&1&\vdots\\
0&\cdots&\cdots&1\\
\end{pmatrix}}_\text{Identity matrix} =
\underbrace{
\begin{pmatrix}
\sigma^2\times \frac{1}{\sqrt{X_1}}&0&\cdots&0\\
0&\sigma^2\times \frac{1}{\sqrt{X_2}}&\cdots&\vdots\\
\vdots&\cdots&\sigma^2\times \frac{1}{\sqrt{X_i}}&\vdots\\
0&\cdots&\cdots&\sigma^2\times \frac{1}{\sqrt{X_n}}\\
\end{pmatrix}}_\text{Variance-covariance matrix}
\]
Dealing with Heterogeneity
\[
\mathbf{V} = \sigma^2\times \omega, \hspace{1cm} \text{where } \omega =
\underbrace{
\begin{pmatrix}
\frac{1}{\sqrt{X_1}}&0&\cdots&0\\
0&\frac{1}{\sqrt{X_2}}&\cdots&\vdots\\
\vdots&\cdots&\frac{1}{\sqrt{X_i}}&\vdots\\
0&\cdots&\cdots&\frac{1}{\sqrt{X_n}}\\
\end{pmatrix}}_\text{Weights matrix}
\]
Dealing with Heterogeneity
Calculating weights
1 /sqrt (data1$x) [1] 1.0000 0.7071 0.5774 0.5000 0.4472 0.4082
[7] 0.3780 0.3536 0.3333 0.3162 0.3015 0.2887
[13] 0.2774 0.2673 0.2582 0.2500
Generalized least squares (GLS)
use OLS to estimate fixed effects
use these estimates to estimate variances via ML
use these to re-estimate fixed effects (OLS)
Generalized least squares (GLS)
ML is biased (for variance) when N is small:
use REML
max. likelihood of residuals rather than data
Variance structures
Variance function
Variance structure
Description
varFixed(~x)
\(V = \sigma^2\times x\)
variance proportional to x the covariate
varExp(form=~x)
\(V = \sigma^2\times e^{2\delta\times x}\)
variance proportional to the exponential of x multiplied by a constant
varPower(form=~x)
\(V = \sigma^2\times |x|^{2\delta}\)
variance proportional to the absolute value of x raised to a constant power
varConstPower(form=~x)
\(V = \sigma^2\times (\delta_1 + |x|^{\delta_2})^2\)
a variant on the power function
varIdent(form=~|A)
\(V = \sigma_j^2\times I\)
when A is a factor, variance is allowed to be different for each level (\(j\)) of the factor
varComb(form=~x|A)
\(V = \sigma_j^2\times x\)
combination of two of the above
Generalized least squares (GLS)
library (nlme)
data1.gls <- gls (y~x, data1,
method= 'REML' )
plot (data1.gls)
library (nlme)
data1.gls1 <- gls (y~x, data= data1, weights= varFixed (~x),
method= 'REML' )
plot (data1.gls1)
Generalized least squares (GLS)
wrong
plot (resid (data1.gls) ~
fitted (data1.gls))
plot (resid (data1.gls1) ~
fitted (data1.gls1))
Generalized least squares (GLS)
CORRECT
plot (resid (data1.gls,'normalized' ) ~
fitted (data1.gls))
plot (resid (data1.gls1,'normalized' ) ~
fitted (data1.gls1))
Generalized least squares (GLS)
plot (resid (data1.gls,'normalized' ) ~ data1$x)
plot (resid (data1.gls1,'normalized' ) ~ data1$x)
Generalized least squares (GLS)
AIC (data1.gls, data1.gls1) df AIC
data1.gls 3 127.6
data1.gls1 3 121.1#OR
anova (data1.gls, data1.gls1) Model df AIC BIC logLik
data1.gls 1 3 127.6 129.6 -60.82
data1.gls1 2 3 121.1 123.0 -57.54
Generalized least squares (GLS)
Can we go better
library (nlme)
data1.gls1 <- gls (y~x, data= data1,
weights= varFixed (~x),method= 'REML' )
plot (data1.gls1)
library (nlme)
data1.gls2 <- gls (y~x, data= data1,
weights= varFixed (~x^2 ),method= 'REML' )
plot (data1.gls2)
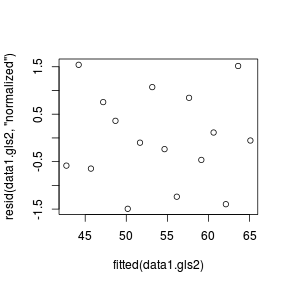
Generalized least squares (GLS)
CORRECT
plot (resid (data1.gls1,'normalized' ) ~
fitted (data1.gls1))
plot (resid (data1.gls2,'normalized' ) ~
fitted (data1.gls2))
Generalized least squares (GLS)
AIC (data1.gls, data1.gls1, data1.gls2) df AIC
data1.gls 3 127.6
data1.gls1 3 121.1
data1.gls2 3 119.0#OR
anova (data1.gls, data1.gls1, data1.gls2) Model df AIC BIC logLik
data1.gls 1 3 127.6 129.6 -60.82
data1.gls1 2 3 121.1 123.0 -57.54
data1.gls2 3 3 119.0 120.9 -56.50
Generalized least squares (GLS)
summary (data1.gls)Generalized least squares fit by REML
Model: y ~ x
Data: data1
AIC BIC logLik
127.6 129.6 -60.82
Coefficients:
Value Std.Error t-value p-value
(Intercept) 40.33 7.189 5.610 0.0001
x 1.57 0.744 2.113 0.0531
Correlation:
(Intr)
x -0.879
Standardized residuals:
Min Q1 Med Q3 Max
-2.00006 -0.29320 -0.02283 0.35358 2.29100
Residual standard error: 13.71
Degrees of freedom: 16 total; 14 residual
summary (data1.gls2)Generalized least squares fit by REML
Model: y ~ x
Data: data1
AIC BIC logLik
119 120.9 -56.5
Variance function:
Structure: fixed weights
Formula: ~x^2
Coefficients:
Value Std.Error t-value p-value
(Intercept) 41.22 1.494 27.598 0.0000
x 1.49 0.470 3.176 0.0067
Correlation:
(Intr)
x -0.671
Standardized residuals:
Min Q1 Med Q3 Max
-1.49260 -0.59853 -0.07669 0.77799 1.54158
Residual standard error: 1.393
Degrees of freedom: 16 total; 14 residual
Generalized least squares (GLS)
data1$cx <- scale (data1$x, scale= FALSE )
data1.gls <- gls (y~cx, data1,
method= 'REML' )
summary (data1.gls)Generalized least squares fit by REML
Model: y ~ cx
Data: data1
AIC BIC logLik
127.6 129.6 -60.82
Coefficients:
Value Std.Error t-value p-value
(Intercept) 53.68 3.427 15.662 0.0000
cx 1.57 0.744 2.113 0.0531
Correlation:
(Intr)
cx 0
Standardized residuals:
Min Q1 Med Q3 Max
-2.00006 -0.29320 -0.02283 0.35358 2.29100
Residual standard error: 13.71
Degrees of freedom: 16 total; 14 residual
data1$cx <- scale (data1$x, scale= FALSE )
data1.gls2 <- gls (y~cx, data1,
weights= varFixed (~x^2 ), method= 'REML' )
summary (data1.gls2)Generalized least squares fit by REML
Model: y ~ cx
Data: data1
AIC BIC logLik
119 120.9 -56.5
Variance function:
Structure: fixed weights
Formula: ~x^2
Coefficients:
Value Std.Error t-value p-value
(Intercept) 53.91 3.19 16.898 0.0000
cx 1.49 0.47 3.176 0.0067
Correlation:
(Intr)
cx 0.938
Standardized residuals:
Min Q1 Med Q3 Max
-1.49260 -0.59853 -0.07669 0.77799 1.54158
Residual standard error: 1.393
Degrees of freedom: 16 total; 14 residual
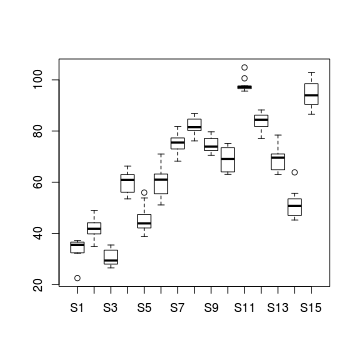
Heteroscadacity in ANOVA
data2 <- read.csv ('../data/D2.csv' )
summary (data2) y x
Min. :29.3 A:10
1st Qu.:36.2 B:10
Median :40.9 C:10
Mean :42.3 D:10
3rd Qu.:49.3 E:10
Max. :56.8
\[
\begin{align*}
y_{i} &= \mu + \alpha_i + \varepsilon_i\\
\epsilon_i&\sim\mathcal{N}(0, \sigma^2) \\
\end{align*}
\]
estimate \(\mu\), \(\alpha_i\) and \(\sigma^2\)
Heteroscadacity in ANOVA
boxplot (y~x, data2)
Heteroscadacity in ANOVA
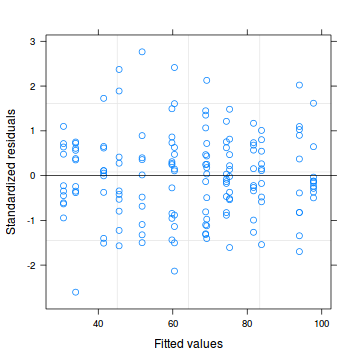
plot (lm (y~x, data2), which= 3 )
Heteroscadacity in ANOVA
\[\varepsilon \sim \mathcal{N}(0,\sigma_i^2\times \omega)\]
\[\text{Effect (}\alpha\text{) 1 (i=1)} \quad \begin{pmatrix}
y_{1i}\\
y_{2i}\\
y_{3i}\\
\end{pmatrix} =
\begin{pmatrix}
1&1&0&0\\
1&1&0&0\\
1&1&0&0\\
\end{pmatrix} \times
\begin{pmatrix}
\beta_i
\end{pmatrix} +
\begin{pmatrix}
\varepsilon_{1i}\\
\varepsilon_{2i}\\
\varepsilon_{3i}
\end{pmatrix}
\quad
\varepsilon_i \sim \mathcal{N}(0,
\begin{pmatrix}
\sigma_1^2&0&0\\
0&\sigma_1^2&0\\
0&0&\sigma_1^2
\end{pmatrix}
)
\] \[\text{Effect (}\alpha\text{) 2 (i=2)} \quad \begin{pmatrix}
y_{1i}\\
y_{2i}\\
y_{3i}\\
\end{pmatrix} =
\begin{pmatrix}
1&0&1&0\\
1&0&1&0\\
1&0&1&0\\
\end{pmatrix} \times
\begin{pmatrix}
\beta_i
\end{pmatrix} +
\begin{pmatrix}
\varepsilon_{1i}\\
\varepsilon_{2i}\\
\varepsilon_{3i}
\end{pmatrix}
\quad
\varepsilon_i \sim \mathcal{N}(0,
\begin{pmatrix}
\sigma_2^2&0&0\\
0&\sigma_2^2&0\\
0&0&\sigma_2^2
\end{pmatrix}
)
\] \[\text{Effect (}\alpha\text{) 3 (i=3)} \quad \begin{pmatrix}
y_{1i}\\
y_{2i}\\
y_{3i}\\
\end{pmatrix} =
\begin{pmatrix}
1&0&0&1\\
1&0&0&1\\
1&0&0&1\\
\end{pmatrix} \times
\begin{pmatrix}
\beta_i
\end{pmatrix} +
\begin{pmatrix}
\varepsilon_{1i}\\
\varepsilon_{2i}\\
\varepsilon_{3i}
\end{pmatrix}
\quad
\varepsilon_i \sim \mathcal{N}(0,
\begin{pmatrix}
\sigma_3^2&0&0\\
0&\sigma_3^2&0\\
0&0&\sigma_3^2
\end{pmatrix}
)
\]
Heteroscadacity in ANOVA
\[ V_{ji} = \begin{pmatrix}
\sigma_1^2&0&0&0&0&0&0&0&0\\
0&\sigma_1^2&0&0&0&0&0&0&0\\
0&0&\sigma_1^2&0&0&0&0&0&0\\
0&0&0&\sigma_2^2&0&0&0&0&0\\
0&0&0&0&\sigma_2^2&0&0&0&0\\
0&0&0&0&0&\sigma_2^2&0&0&0\\
0&0&0&0&0&0&\sigma_3^2&0&0\\
0&0&0&0&0&0&0&\sigma_3^2&0\\
0&0&0&0&0&0&0&0&\sigma_3^2\\
\end{pmatrix}
\]
Heteroscadacity in ANOVA
data2.sd <- with (data2, tapply (y,x,sd))
1 /(data2.sd[1 ]/data2.sd) A B C D E
1.00000 0.91343 0.40807 0.08632 0.12720
Variance structures
Variance function
Variance structure
Description
varFixed(~x)
\(V = \sigma^2\times x\)
variance proportional to x the covariate
varExp(form=~x)
\(V = \sigma^2\times e^{2\delta\times x}\)
variance proportional to the exponential of x multiplied by a constant
varPower(form=~x)
\(V = \sigma^2\times |x|^{2\delta}\)
variance proportional to the absolute value of x raised to a constant power
varConstPower(form=~x)
\(V = \sigma^2\times (\delta_1 + |x|^{\delta_2})^2\)
a variant on the power function
varIdent(form=~|A)
\(V = \sigma_j^2\times I\)
when A is a factor, variance is allowed to be different for each level (\(j\)) of the factor
varComb(form=~x|A)
\(V = \sigma_j^2\times x\)
combination of two of the above
Heteroscadacity in ANOVA
library (nlme)
data2.gls <- gls (y~x, data= data2,
method= "REML" )
library (nlme)
data2.gls1 <- gls (y~x, data= data2,
weights= varIdent (form= ~1 |x), method= "REML" )
Heteroscadacity in ANOVA
library (nlme)
data2.gls <- gls (y~x, data= data2,
method= "REML" )
plot (resid (data2.gls,'normalized' ) ~
fitted (data2.gls))
library (nlme)
data2.gls1 <- gls (y~x, data= data2,
weights= varIdent (form= ~1 |x), method= "REML" )
plot (resid (data2.gls1,'normalized' ) ~
fitted (data2.gls1))
Heteroscadacity in ANOVA
AIC (data2.gls,data2.gls1) df AIC
data2.gls 6 249.5
data2.gls1 10 199.2anova (data2.gls,data2.gls1) Model df AIC BIC logLik Test
data2.gls 1 6 249.5 260.3 -118.8
data2.gls1 2 10 199.2 217.3 -89.6 1 vs 2
L.Ratio p-value
data2.gls
data2.gls1 58.29 <.0001
Heteroscadacity in ANOVA
summary (data2.gls)Generalized least squares fit by REML
Model: y ~ x
Data: data2
AIC BIC logLik
249.5 260.3 -118.7
Coefficients:
Value Std.Error t-value p-value
(Intercept) 40.79 0.9424 43.29 0.0000
xB 5.20 1.3328 3.90 0.0003
xC 13.94 1.3328 10.46 0.0000
xD -0.73 1.3328 -0.55 0.5851
xE -10.66 1.3328 -8.00 0.0000
Correlation:
(Intr) xB xC xD
xB -0.707
xC -0.707 0.500
xD -0.707 0.500 0.500
xE -0.707 0.500 0.500 0.500
Standardized residuals:
Min Q1 Med Q3 Max
-3.30654 -0.24626 0.06468 0.39047 2.94559
Residual standard error: 2.98
Degrees of freedom: 50 total; 45 residual
summary (data2.gls1)Generalized least squares fit by REML
Model: y ~ x
Data: data2
AIC BIC logLik
199.2 217.3 -89.6
Variance function:
Structure: Different standard deviations per stratum
Formula: ~1 | x
Parameter estimates:
A B C D E
1.00000 0.91342 0.40807 0.08632 0.12720
Coefficients:
Value Std.Error t-value p-value
(Intercept) 40.79 1.481 27.543 0.0000
xB 5.20 2.006 2.593 0.0128
xC 13.94 1.600 8.714 0.0000
xD -0.73 1.487 -0.493 0.6244
xE -10.66 1.493 -7.139 0.0000
Correlation:
(Intr) xB xC xD
xB -0.738
xC -0.926 0.684
xD -0.996 0.736 0.922
xE -0.992 0.732 0.918 0.988
Standardized residuals:
Min Q1 Med Q3 Max
-2.3034 -0.6179 0.1065 0.7597 1.8743
Residual standard error: 4.684
Degrees of freedom: 50 total; 45 residual
Bayesian
library (R2jags)
modelString = "
model {
#Likelihood
for (i in 1:n) {
y[i]~dnorm(mu[i],tau)
mu[i] <- beta0+beta1*x[i]
}
#Priors
beta0 ~ dnorm(0.01,1.0E-6)
beta1 ~ dnorm(0,1.0E-6)
tau <- 1 / (sigma*sigma)
sigma~dunif(0,100)
}
"
writeLines (modelString, con= '../BUGSscripts/simpleRegression.JAGS' )
data1.list <- list (y = data1$y, x = data1$x,
n = nrow (data1))
params <- c ("beta0" , "beta1" , "sigma" )
nChains = 3
burnInSteps = 3000
thinSteps = 10
numSavedSteps = 15000
nIter = ceiling (burnInSteps+(numSavedSteps * thinSteps)/nChains)
nIter[1] 53000data1.r2jags <- jags (data= data1.list,
inits= NULL ,
parameters.to.save= params,
model.file= "../BUGSscripts/simpleRegression.JAGS" ,
n.chains= nChains,
n.iter= nIter,
n.burnin= burnInSteps,
n.thin= thinSteps
) Compiling model graph
Resolving undeclared variables
Allocating nodes
Graph Size: 121
Initializing modelprint (data1.r2jags) Inference for Bugs model at "../BUGSscripts/simpleRegression.JAGS", fit using jags,
3 chains, each with 53000 iterations (first 3000 discarded), n.thin = 10
n.sims = 15000 iterations saved
mu.vect sd.vect 2.5% 25% 50%
beta0 50.503 2.642 45.318 48.742 50.509
beta1 -2.716 0.799 -4.283 -3.259 -2.707
sigma 7.965 0.844 6.540 7.377 7.889
deviance 347.934 2.585 345.008 346.051 347.273
75% 97.5% Rhat n.eff
beta0 52.251 55.705 1.001 15000
beta1 -2.185 -1.154 1.001 15000
sigma 8.487 9.856 1.001 15000
deviance 349.079 354.550 1.001 3700
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 3.3 and DIC = 351.3
DIC is an estimate of expected predictive error (lower deviance is better).summary (data1.gls)Generalized least squares fit by REML
Model: y ~ cx
Data: data1
AIC BIC logLik
127.6 129.6 -60.82
Coefficients:
Value Std.Error t-value p-value
(Intercept) 53.68 3.427 15.662 0.0000
cx 1.57 0.744 2.113 0.0531
Correlation:
(Intr)
cx 0
Standardized residuals:
Min Q1 Med Q3 Max
-2.00006 -0.29320 -0.02283 0.35358 2.29100
Residual standard error: 13.71
Degrees of freedom: 16 total; 14 residualdata1.gls$sigma[1] 13.71modelString = "
model {
#Likelihood
for (i in 1:n) {
y[i]~dnorm(mu[i],tau*(1/x[i]))
mu[i] <- beta0+beta1*x[i]
}
#Priors
beta0 ~ dnorm(0.01,1.0E-6)
beta1 ~ dnorm(0,1.0E-6)
tau <- 1 / (sigma*sigma)
sigma~dunif(0,100)
}
"
writeLines (modelString, con= '../BUGSscripts/simpleRegression1.JAGS' )
data1.list <- with (data1, list (y = y, x = x, n = nrow (data1)))
params <- c ("beta0" , "beta1" , "sigma" )
nChains = 3
burnInSteps = 3000
thinSteps = 10
numSavedSteps = 15000
nIter = ceiling (burnInSteps+(numSavedSteps * thinSteps)/nChains)
nIter[1] 53000data1.r2jags1 <- jags (data= data1.list,
inits= NULL ,
parameters.to.save= params,
model.file= "../BUGSscripts/simpleRegression1.JAGS" ,
n.chains= nChains,
n.iter= nIter,
n.burnin= burnInSteps,
n.thin= thinSteps
) Compiling model graph
Resolving undeclared variables
Allocating nodes
Graph Size: 131
Initializing modelprint (data1.r2jags1) Inference for Bugs model at "../BUGSscripts/simpleRegression1.JAGS", fit using jags,
3 chains, each with 53000 iterations (first 3000 discarded), n.thin = 10
n.sims = 15000 iterations saved
mu.vect sd.vect 2.5% 25% 50%
beta0 44.620 2.047 40.624 43.260 44.609
beta1 -0.756 0.797 -2.317 -1.288 -0.756
sigma 5.080 0.535 4.156 4.703 5.032
deviance 350.703 2.555 347.786 348.829 350.061
75% 97.5% Rhat n.eff
beta0 46.001 48.639 1.001 15000
beta1 -0.222 0.803 1.001 15000
sigma 5.407 6.253 1.001 12000
deviance 351.875 357.345 1.001 15000
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 3.3 and DIC = 354.0
DIC is an estimate of expected predictive error (lower deviance is better).summary (data1.gls1)Generalized least squares fit by REML
Model: y ~ x
Data: data1
AIC BIC logLik
121.1 123 -57.54
Variance function:
Structure: fixed weights
Formula: ~x
Coefficients:
Value Std.Error t-value p-value
(Intercept) 41.50 3.332 12.455 0.0000
x 1.43 0.525 2.727 0.0164
Correlation:
(Intr)
x -0.746
Standardized residuals:
Min Q1 Med Q3 Max
-1.74684 -0.42653 -0.05467 0.56196 2.04503
Residual standard error: 4.079
Degrees of freedom: 16 total; 14 residualdata1.gls1$sigma[1] 4.079modelString = "
model {
#Likelihood
for (i in 1:n) {
y[i]~dnorm(mu[i],tau*(1/x[i]^2))
mu[i] <- beta0+beta1*x[i]
}
#Priors
beta0 ~ dnorm(0.01,1.0E-6)
beta1 ~ dnorm(0,1.0E-6)
tau <- 1 / (sigma*sigma)
sigma~dunif(0,100)
}
"
writeLines (modelString, con= '../BUGSscripts/simpleRegression2.JAGS' )
data1.list <- with (data1, list (y = y, x = x, n = nrow (data1)))
params <- c ("beta0" , "beta1" , "sigma" )
nChains = 3
burnInSteps = 3000
thinSteps = 10
numSavedSteps = 15000
nIter = ceiling (burnInSteps+(numSavedSteps * thinSteps)/nChains)
nIter[1] 53000data1.r2jags2 <- jags (data= data1.list,
inits= NULL ,
parameters.to.save= params,
model.file= "../BUGSscripts/simpleRegression2.JAGS" ,
n.chains= nChains,
n.iter= nIter,
n.burnin= burnInSteps,
n.thin= thinSteps
) Compiling model graph
Resolving undeclared variables
Allocating nodes
Graph Size: 137
Initializing modelprint (data1.r2jags2) Inference for Bugs model at "../BUGSscripts/simpleRegression2.JAGS", fit using jags,
3 chains, each with 53000 iterations (first 3000 discarded), n.thin = 10
n.sims = 15000 iterations saved
mu.vect sd.vect 2.5% 25% 50%
beta0 41.087 1.666 37.792 39.985 41.076
beta1 0.846 0.900 -0.915 0.251 0.844
sigma 3.390 0.357 2.772 3.135 3.363
deviance 358.160 2.585 355.239 356.287 357.494
75% 97.5% Rhat n.eff
beta0 42.195 44.356 1.001 14000
beta1 1.442 2.618 1.001 15000
sigma 3.612 4.174 1.001 15000
deviance 359.321 364.845 1.001 15000
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 3.3 and DIC = 361.5
DIC is an estimate of expected predictive error (lower deviance is better).summary (data1.gls2)Generalized least squares fit by REML
Model: y ~ cx
Data: data1
AIC BIC logLik
119 120.9 -56.5
Variance function:
Structure: fixed weights
Formula: ~x^2
Coefficients:
Value Std.Error t-value p-value
(Intercept) 53.91 3.19 16.898 0.0000
cx 1.49 0.47 3.176 0.0067
Correlation:
(Intr)
cx 0.938
Standardized residuals:
Min Q1 Med Q3 Max
-1.49260 -0.59853 -0.07669 0.77799 1.54158
Residual standard error: 1.393
Degrees of freedom: 16 total; 14 residualdata1.gls2$sigma[1] 1.393modelString="
model {
#Likelihood
for (i in 1:n) {
y[i]~dnorm(mean[i],tau)
mean[i] <- X[i,] %*% beta
}
#Priors and derivatives
for (i in 1:ngroups) {
beta[i] ~ dnorm(0, 1.0E-6)
}
sigma ~ dunif(0, 1000)
tau <- 1 / (sigma * sigma)
}
"
writeLines (modelString,con= "../BUGSscripts/simpleANOVA.JAGS" )
X <- model.matrix (~x, data2)
data2.list <- list (y= data2$y, X= X, n= nrow (data2), ngroups= ncol (X))
params <- c ("beta" ,"sigma" )
burnInSteps = 2000
nChains = 3
numSavedSteps = 50000
thinSteps = 10
nIter = ceiling ((numSavedSteps * thinSteps)/nChains)
library (R2jags)
data2.r2jags <- jags (data= data2.list,
inits= NULL ,
parameters.to.save= params,
model.file= "../BUGSscripts/simpleANOVA.JAGS" ,
n.chains= 3 ,
n.iter= nIter,
n.burnin= burnInSteps,
n.thin= thinSteps
)Compiling model graph
Resolving undeclared variables
Allocating nodes
Graph Size: 370
Initializing modelprint (data2.r2jags)Inference for Bugs model at "../BUGSscripts/simpleANOVA.JAGS", fit using jags,
3 chains, each with 166667 iterations (first 2000 discarded), n.thin = 10
n.sims = 49401 iterations saved
mu.vect sd.vect 2.5% 25% 50%
beta[1] 40.800 0.976 38.869 40.154 40.806
beta[2] 5.188 1.379 2.495 4.265 5.184
beta[3] 13.930 1.380 11.229 13.018 13.927
beta[4] -0.744 1.379 -3.433 -1.661 -0.746
beta[5] -10.662 1.378 -13.367 -11.581 -10.664
sigma 3.068 0.334 2.498 2.832 3.039
deviance 252.356 3.766 247.195 249.599 251.639
75% 97.5% Rhat n.eff
beta[1] 41.451 42.707 1.001 49000
beta[2] 6.099 7.906 1.001 49000
beta[3] 14.843 16.655 1.001 49000
beta[4] 0.171 1.986 1.001 49000
beta[5] -9.751 -7.946 1.001 49000
sigma 3.271 3.803 1.001 40000
deviance 254.374 261.631 1.001 49000
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 7.1 and DIC = 259.4
DIC is an estimate of expected predictive error (lower deviance is better).summary (data2.gls)Generalized least squares fit by REML
Model: y ~ x
Data: data2
AIC BIC logLik
249.5 260.3 -118.7
Coefficients:
Value Std.Error t-value p-value
(Intercept) 40.79 0.9424 43.29 0.0000
xB 5.20 1.3328 3.90 0.0003
xC 13.94 1.3328 10.46 0.0000
xD -0.73 1.3328 -0.55 0.5851
xE -10.66 1.3328 -8.00 0.0000
Correlation:
(Intr) xB xC xD
xB -0.707
xC -0.707 0.500
xD -0.707 0.500 0.500
xE -0.707 0.500 0.500 0.500
Standardized residuals:
Min Q1 Med Q3 Max
-3.30654 -0.24626 0.06468 0.39047 2.94559
Residual standard error: 2.98
Degrees of freedom: 50 total; 45 residualdata2$sigmaNULLmodelString="
model {
#Likelihood
for (i in 1:n) {
y[i]~dnorm(mean[i],tau[x[i]])
mean[i] <- X[i,] %*% eta
}
#Priors and derivatives
for (i in 1:ngroups) {
beta[i] ~ dnorm(0, 1.0E-6) #prior
sigma[i] ~ dgamma(0.001, 0.001)
tau[i] <- 1 / (sigma[i] * sigma[i])
}
}
"
writeLines (modelString,con= "../BUGSscripts/simpleANOVA1.JAGS" )
X <- model.matrix (~x, data2)
data2.list <- list (y= data2$y, X= X, x= as.numeric (data2$x), n= nrow (data2), ngroups= ncol (X))
params <- c ("beta" ,"sigma" )
burnInSteps = 2000
nChains = 3
numSavedSteps = 50000
thinSteps = 10
nIter = ceiling ((numSavedSteps * thinSteps)/nChains)
library (R2jags)
data2.r2jags1 <- jags (data= data2.list,
inits= NULL ,
parameters.to.save= params,
model.file= "../BUGSscripts/simpleANOVA1.JAGS" ,
n.chains= 3 ,
n.iter= nIter,
n.burnin= burnInSteps,
n.thin= thinSteps
)Compiling model graph
Resolving undeclared variables
Allocating nodes
Deleting modelError: RUNTIME ERROR:
Compilation error on line 5.
Unable to resolve node mean[1]
This may be due to an undefined ancestor node or a directed cycle in the graphprint (data2.r2jags1)Error: object 'data2.r2jags1' not foundsummary (data2.gls1)Generalized least squares fit by REML
Model: y ~ x
Data: data2
AIC BIC logLik
199.2 217.3 -89.6
Variance function:
Structure: Different standard deviations per stratum
Formula: ~1 | x
Parameter estimates:
A B C D E
1.00000 0.91342 0.40807 0.08632 0.12720
Coefficients:
Value Std.Error t-value p-value
(Intercept) 40.79 1.481 27.543 0.0000
xB 5.20 2.006 2.593 0.0128
xC 13.94 1.600 8.714 0.0000
xD -0.73 1.487 -0.493 0.6244
xE -10.66 1.493 -7.139 0.0000
Correlation:
(Intr) xB xC xD
xB -0.738
xC -0.926 0.684
xD -0.996 0.736 0.922
xE -0.992 0.732 0.918 0.988
Standardized residuals:
Min Q1 Med Q3 Max
-2.3034 -0.6179 0.1065 0.7597 1.8743
Residual standard error: 4.684
Degrees of freedom: 50 total; 45 residualdata2.gls1$sigma[1] 4.684
Linear modelling assumptions
\[
\begin{align*}
y_{i} &= \beta_0 + \beta_1 \times x_{i} + \varepsilon_i\\
\epsilon_i&\sim\mathcal{N}(0, \sigma^2) \\
\end{align*}
\]
Non-independence
one response is triggered by another
temporal/spatial autocorrelation nested (hierarchical) design structures
Variance-covariance
\[
\mathbf{V} = \underbrace{
\begin{pmatrix}
\sigma^2&0&\cdots&0\\
0&\sigma^2&\cdots&\vdots\\
\vdots&\cdots&\sigma^2&\vdots\\
0&\cdots&\cdots&\sigma^2\\
\end{pmatrix}}_\text{Variance-covariance matrix}
\]
Compound symmetry
constant correlation (and cov)
sphericity
\[
cor(\varepsilon)= \underbrace{
\begin{pmatrix}
1&\rho&\cdots&\rho\\
\rho&1&\cdots&\vdots\\
\dots&\cdots&1&\vdots\\
\rho&\cdots&\cdots&1\\
\end{pmatrix}}_\text{Correlation matrix}
\] \[
\mathbf{V} = \underbrace{
\begin{pmatrix}
\theta + \sigma^2&\theta&\cdots&\theta\\
\theta&\theta + \sigma^2&\cdots&\vdots\\
\vdots&\cdots&\theta + \sigma^2&\vdots\\
\theta&\cdots&\cdots&\theta + \sigma^2\\
\end{pmatrix}}_\text{Variance-covariance matrix}
\]
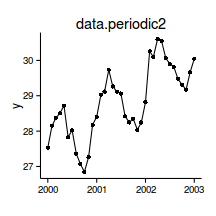
Temporal autocorrelation
correlation dependent on proximity
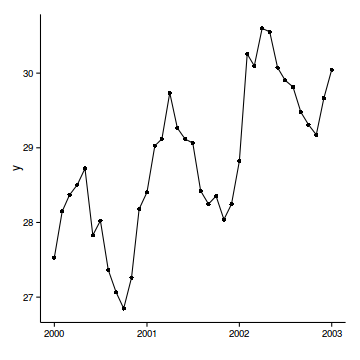
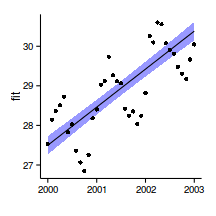
data.t
Temporal autocorrelation
Relationship between Y and X
data.t.lm <- lm (y~x, data= data.t)
par (mfrow= c (2 ,3 ))
plot (data.t.lm, which= 1 :6 , ask= FALSE )
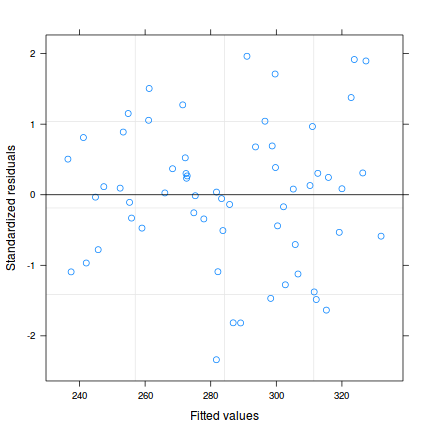
Temporal autocorrelation
Relationship between Y and X
acf (rstandard (data.t.lm))
Temporal autocorrelation
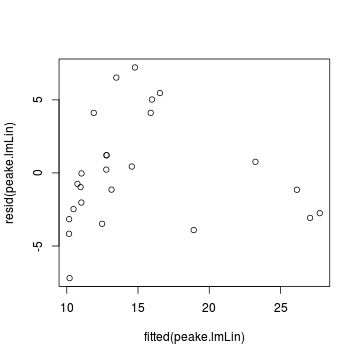
plot (rstandard (data.t.lm)~data.t$year)
Temporal autocorrelation
library (car)
vif (lm (y~x+year, data= data.t)) x year
1.04 1.04 data.t.lm1 <- lm (y~x+year, data.t)
Testing for autocorrelation
Residual plot
par (mfrow= c (1 ,2 ))
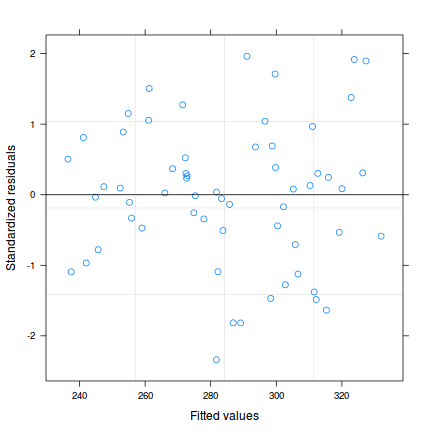
plot (rstandard (data.t.lm1)~fitted (data.t.lm1))
plot (rstandard (data.t.lm1)~data.t$year)
Testing for autocorrelation
Autocorrelation (acf) plot
acf (rstandard (data.t.lm1), lag= 40 )
First order autocorrelation (AR1)
\[
cor(\varepsilon) = \underbrace{
\begin{pmatrix}
1&\rho&\cdots&\rho^{|t-s|}\\
\rho&1&\cdots&\vdots\\
\vdots&\cdots&1&\vdots\\
\rho^{|t-s|}&\cdots&\cdots&1\\
\end{pmatrix}}_\text{First order autoregressive correlation structure}
\]
where:
\(s\) and \(t\) are the times.
\(s-t\) is the lag
First order auto-regressive (AR1)
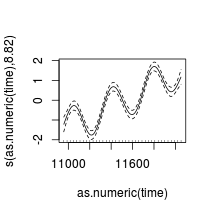
library (nlme)
data.t.gls <- gls (y~x+year, data= data.t, method= 'REML' )
data.t.gls1 <- gls (y~x+year, data= data.t,
correlation= corAR1 (form= ~year),method= 'REML' )
First order auto-regressive (AR1)
par (mfrow= c (1 ,2 ))
plot (residuals (data.t.gls, type= "normalized" )~
fitted (data.t.gls))
plot (residuals (data.t.gls1, type= "normalized" )~
fitted (data.t.gls1))
First order auto-regressive (AR1)
par (mfrow= c (1 ,2 ))
plot (residuals (data.t.gls, type= "normalized" )~
data.t$x)
plot (residuals (data.t.gls1, type= "normalized" )~
data.t$x)
First order auto-regressive (AR1)
par (mfrow= c (1 ,2 ))
plot (residuals (data.t.gls, type= "normalized" )~
data.t$year)
plot (residuals (data.t.gls1, type= "normalized" )~
data.t$year)
First order auto-regressive (AR1)
par (mfrow= c (1 ,2 ))
acf (residuals (data.t.gls, type= 'normalized' ), lag= 40 )
acf (residuals (data.t.gls1, type= 'normalized' ), lag= 40 )
First order auto-regressive (AR1)
AIC (data.t.gls, data.t.gls1) df AIC
data.t.gls 4 626.3
data.t.gls1 5 536.7anova (data.t.gls, data.t.gls1) Model df AIC BIC logLik Test
data.t.gls 1 4 626.3 636.6 -309.2
data.t.gls1 2 5 536.7 549.6 -263.4 1 vs 2
L.Ratio p-value
data.t.gls
data.t.gls1 91.58 <.0001
Auto-regressive moving average (ARMA)
data.t.gls2 <- update (data.t.gls,
correlation= corARMA (form= ~year,p= 2 ,q= 0 ))
data.t.gls3 <- update (data.t.gls,
correlation= corARMA (form= ~year,p= 3 ,q= 0 ))
AIC (data.t.gls, data.t.gls1, data.t.gls2, data.t.gls3) df AIC
data.t.gls 4 626.3
data.t.gls1 5 536.7
data.t.gls2 6 538.1
data.t.gls3 7 538.8
Summarize model
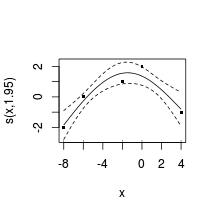
summary (data.t.gls1)Generalized least squares fit by REML
Model: y ~ x + year
Data: data.t
AIC BIC logLik
536.7 549.6 -263.4
Correlation Structure: ARMA(1,0)
Formula: ~year
Parameter estimate(s):
Phi1
0.9127
Coefficients:
Value Std.Error t-value p-value
(Intercept) 4389 1232.6 3.560 0.0006
x 0 0.0 3.297 0.0014
year -2 0.6 -3.546 0.0006
Correlation:
(Intr) x
x 0.009
year -1.000 -0.010
Standardized residuals:
Min Q1 Med Q3 Max
-1.423 1.711 3.378 4.373 6.624
Residual standard error: 3.375
Degrees of freedom: 100 total; 97 residual
Spatial autocorrelation
\[
cor(\varepsilon) = \underbrace{
\begin{pmatrix}
1&e^{-\delta}&\cdots&e^{-\delta D}\\
e^{-\delta}&1&\cdots&\vdots\\
\vdots&\cdots&1&\vdots\\
e^{-\delta D}&\cdots&\cdots&1\\
\end{pmatrix}}_\text{Exponential autoregressive correlation structure}
\]
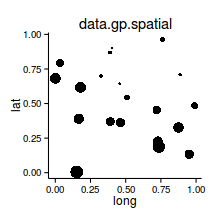
Spatial autocorrelation
Spatial autocorrelation
library (sp)
coordinates (data.s) <- ~LAT+LONG
bubble (data.s,'y' )
Spatial autocorrelation
Relationship between Y and X
data.s.lm <- lm (y~x, data= data.s)
par (mfrow= c (2 ,3 ))
plot (data.s.lm, which= 1 :6 , ask= FALSE )
Detecting spatial autocorrelation
bubble plot
semi-variogram
Bubble plot
data.s$Resid <- rstandard (data.s.lm)
library (sp)
coordinates (data.s) <- ~LAT+LONGError: setting coordinates cannot be done on
Spatial objects, where they have already been setbubble (data.s,'Resid' )
Semi-variogram
semivariance = similarity (of residuals) between pairs at specific distancesdistances binned according to distance and orientation (N)
Semi-variogram
Semi-variogram
library (nlme)
data.s.gls <- gls (y~x, data.s, method= 'REML' )
plot (nlme:::Variogram (data.s.gls, form= ~LAT+LONG,
resType= "normalized" ))
Semi-variogram
library (gstat)
plot (variogram (residuals (data.s.gls,"normalized" )~1 ,
data= data.s, cutoff= 6 ))
Semi-variogram
library (gstat)
plot (variogram (residuals (data.s.gls,"normalized" )~1 ,
data= data.s, cutoff= 6 ,alpha= c (0 ,45 ,90 ,135 )))
Accommodating spatial autocorrelation
correlation function
Correlation structure
Description
corExp(form=~lat+long)
Exponential
\(\Phi = 1-e^{-D/\rho}\)
corGaus(form=~lat+long)
Gaussian
\(\Phi = 1-e^{-(D/\rho)^2}\)
corLin(form=~lat+long)
Linear
\(\Phi = 1-(1-D/\rho)I(d<\rho)\)
corRatio(form=~lat+long)
Rational quadratic
\(\Phi = (d/\rho)^2/(1+(d/\rho)2)\)
corSpher(form=~lat+long)
Spherical
\(\Phi = 1-(1-1.5(d/\rho) + 0.5(d/\rho)^3)I(d<\rho)\)
Accommodating spatial autocorrelation
data.s.glsExp <- update (data.s.gls,
correlation= corExp (form= ~LAT+LONG, nugget= TRUE ))
data.s.glsGaus <- update (data.s.gls,
correlation= corGaus (form= ~LAT+LONG, nugget= TRUE ))
#data.s.glsLin <- update(data.s/gls,
# correlation=corLin(form=~LAT+LONG, nugget=TRUE))
data.s.glsRatio <- update (data.s.gls,
correlation= corRatio (form= ~LAT+LONG, nugget= TRUE ))
data.s.glsSpher <- update (data.s.gls,
correlation= corSpher (form= ~LAT+LONG, nugget= TRUE ))
AIC (data.s.gls, data.s.glsExp, data.s.glsGaus, data.s.glsRatio, data.s.glsSpher) df AIC
data.s.gls 3 1013.9
data.s.glsExp 5 974.3
data.s.glsGaus 5 976.4
data.s.glsRatio 5 974.8
data.s.glsSpher 5 975.5
Accommodating spatial autocorrelation
plot (residuals (data.s.glsExp, type= "normalized" )~
fitted (data.s.glsExp))
Accommodating spatial autocorrelation
plot (nlme:::Variogram (data.s.glsExp, form= ~LAT+LONG,
resType= "normalized" ))
Summarize model
summary (data.s.glsExp)Generalized least squares fit by REML
Model: y ~ x
Data: data.s
AIC BIC logLik
974.3 987.2 -482.2
Correlation Structure: Exponential spatial correlation
Formula: ~LAT + LONG
Parameter estimate(s):
range nugget
1.6957 0.1281
Coefficients:
Value Std.Error t-value p-value
(Intercept) 65.90 21.825 3.020 0.0032
x 0.95 0.286 3.304 0.0013
Correlation:
(Intr)
x -0.418
Standardized residuals:
Min Q1 Med Q3 Max
-1.6019 -0.3508 0.1609 0.6452 2.1332
Residual standard error: 47.69
Degrees of freedom: 100 total; 98 residualanova (data.s.glsExp)Denom. DF: 98
numDF F-value p-value
(Intercept) 1 23.46 <.0001
x 1 10.92 0.0013
Summarize model
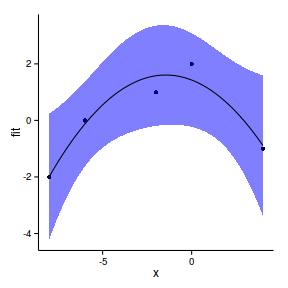
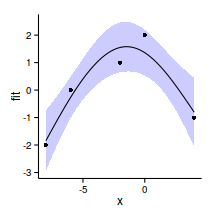
xs <- seq (min (data.s$x), max (data.s$x), l= 100 )
xmat <- model.matrix (~x, data.frame (x= xs))
mpred <- function(model, xmat, data.s) {
pred <- as.vector (coef (model) %*% t (xmat))
(se<-sqrt (diag (xmat %*% vcov (model) %*% t (xmat))))
ci <- data.frame (pred+outer (se,qt (df= nrow (data.s)-2 ,c (.025 ,.975 ))))
colnames (ci) <- c ('lwr' ,'upr' )
data.s.sum<-data.frame (pred, x= xs,se,ci)
data.s.sum
}
data.s.sum<-mpred (data.s.glsExp, xmat, data.s)
plot (pred~x, ylim= c (min (lwr), max (upr)), data.s.sum, type= "l" )
lines (lwr~x, data.s.sum)
lines (upr~x, data.s.sum)
points (pred+residuals (data.s.glsExp)~x, data.s.sum)
newdata <- data.frame (x= xs)[1 ,]
Non-independence
one response is triggered by another
temporal/spatial autocorrelation
nested (hierarchical) design structures