Important data manipulation libraries
| Task | Function | Package |
|---|---|---|
| Sorting | order() |
base |
arrange() |
dplyr | |
| Re-ordering factor levels | factor(,levels=) |
base |
reorder(,new.order=) |
gdata | |
| Re-labelling | factor(,lab=) |
base |
recode() |
dplyr | |
revalue(,replace=) |
plyr | |
| Re-naming columns | colnames() |
base |
rename(,replace=) |
dplyr | |
| Filtering/Subsetting | indexing | base |
subset(,subset=,select=) |
base | |
select(,...) |
dplyr |
Important data manipulation libraries
| Task | Function | Package |
|---|---|---|
| Transformations | transform(), within() |
base |
mutate() |
dplyr | |
| Adding columns | within() |
base |
mutate() |
dplyr | |
| Reshaping data | gather(), spread() |
tidyr |
melt(), cast() |
reshape2 | |
| Aggregating | tapply() |
base |
group_by() |
dplyr | |
cast() |
reshape2 | |
summaryBy() |
doBy | |
| Merging/joining | merge() |
base |
*_join() |
dplyr |
The grammar of data manipulation
Verbs
arrange()- sorting dataselect()- subset columnsrename()- rename columnsfilter()- subset rowsslice()mutate()- adding columssummarise()- aggregate (group_by())count()- tally
The grammar of data manipulation
Tidying verbs
gather()- melt to long formatspread()- cast to wide formatunite()- combine columnsseparate()- separate columns
multi data.frame verbs
*_join()- merging data
The grammar of data manipulation
Piping
%>%
The grammar of data manipulation
https://www.rstudio.com/resources/cheatsheets/data-transformation.pdf
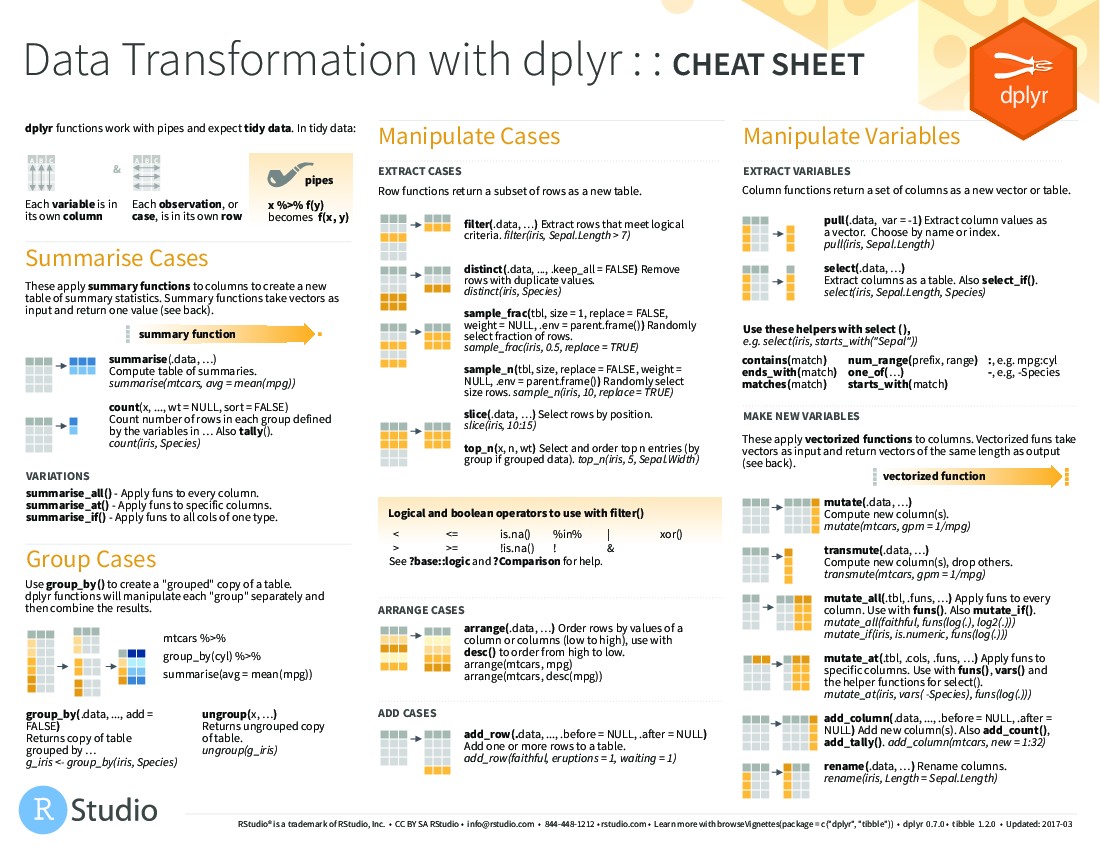
<img src=“figure/data-transformation.png”, width=“550px”>
Data files
| Between | Plot | Cond | Time | Temp | LAT | LONG |
|---|---|---|---|---|---|---|
| A1 | P1 | H | 1 | 15.74 | 17.26 | 146.2 |
| A1 | P1 | M | 2 | 23.84 | 14.07 | 144.9 |
| A1 | P1 | L | 3 | 13.64 | 20.75 | 144.7 |
| A1 | P2 | H | 4 | 37.95 | 18.41 | 142.1 |
| A1 | P2 | M | 1 | 25.3 | 18.47 | 144 |
| A1 | P2 | L | 2 | 13.8 | 20.39 | 145.8 |
| A2 | P3 | H | 3 | 26.87 | 20.14 | 147.7 |
| A2 | P3 | M | 4 | 29.38 | 19.69 | 144.8 |
| A2 | P3 | L | 1 | 27.76 | 20.34 | 145.8 |
| A2 | P4 | H | 2 | 18.95 | 20.06 | 144.9 |
| A2 | P4 | M | 3 | 37.12 | 18.65 | 142.2 |
| A2 | P4 | L | 4 | 25.9 | 14.52 | 144.2 |
Data manipulation packages
Data files
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877
3 A1 P1 L 3 13.64371 20.74986 144.6884
4 A1 P2 H 4 37.95281 18.41013 142.0585
5 A1 P2 M 1 25.29508 18.46762 144.0437
6 A1 P2 L 2 13.79532 20.38767 145.8359 Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877
3 A1 P1 L 3 13.64371 20.74986 144.6884
4 A1 P2 H 4 37.95281 18.41013 142.0585
5 A1 P2 M 1 25.29508 18.46762 144.0437
6 A1 P2 L 2 13.79532 20.38767 145.8359Data files
Between Plot Cond Time Temp LAT LONG
A1:6 P1:3 H:4 Min. :1.00 Min. :13.64 Min. :14.07 Min. :142.1
A2:6 P2:3 L:4 1st Qu.:1.75 1st Qu.:18.14 1st Qu.:18.12 1st Qu.:144.1
P3:3 M:4 Median :2.50 Median :25.60 Median :19.17 Median :144.8
P4:3 Mean :2.50 Mean :24.69 Mean :18.56 Mean :144.8
3rd Qu.:3.25 3rd Qu.:28.16 3rd Qu.:20.19 3rd Qu.:145.8
Max. :4.00 Max. :37.95 Max. :20.75 Max. :147.7 Data files
Between Plot Cond Time Temp LAT LONG
A1:6 P1:3 H:4 Min. :1.00 Min. :13.64 Min. :14.07 Min. :142.1
A2:6 P2:3 L:4 1st Qu.:1.75 1st Qu.:18.14 1st Qu.:18.12 1st Qu.:144.1
P3:3 M:4 Median :2.50 Median :25.60 Median :19.17 Median :144.8
P4:3 Mean :2.50 Mean :24.69 Mean :18.56 Mean :144.8
3rd Qu.:3.25 3rd Qu.:28.16 3rd Qu.:20.19 3rd Qu.:145.8
Max. :4.00 Max. :37.95 Max. :20.75 Max. :147.7 Between Plot Cond Time Temp LAT LONG
A1:6 P1:3 H:4 Min. :1.00 Min. :13.64 Min. :14.07 Min. :142.1
A2:6 P2:3 L:4 1st Qu.:1.75 1st Qu.:18.14 1st Qu.:18.12 1st Qu.:144.1
P3:3 M:4 Median :2.50 Median :25.60 Median :19.17 Median :144.8
P4:3 Mean :2.50 Mean :24.69 Mean :18.56 Mean :144.8
3rd Qu.:3.25 3rd Qu.:28.16 3rd Qu.:20.19 3rd Qu.:145.8
Max. :4.00 Max. :37.95 Max. :20.75 Max. :147.7 Data files
'data.frame': 12 obs. of 7 variables:
$ Between: Factor w/ 2 levels "A1","A2": 1 1 1 1 1 1 2 2 2 2 ...
$ Plot : Factor w/ 4 levels "P1","P2","P3",..: 1 1 1 2 2 2 3 3 3 4 ...
$ Cond : Factor w/ 3 levels "H","L","M": 1 3 2 1 3 2 1 3 2 1 ...
$ Time : int 1 2 3 4 1 2 3 4 1 2 ...
$ Temp : num 15.7 23.8 13.6 38 25.3 ...
$ LAT : num 17.3 14.1 20.7 18.4 18.5 ...
$ LONG : num 146 145 145 142 144 ...
- attr(*, "out.attrs")=List of 2
..$ dim : Named int 3 4
.. ..- attr(*, "names")= chr "Cond" "Plot"
..$ dimnames:List of 2
.. ..$ Cond: chr "Cond=H" "Cond=M" "Cond=L"
.. ..$ Plot: chr "Plot=P1" "Plot=P2" "Plot=P3" "Plot=P4"Dense summary
Observations: 12
Variables: 7
$ Between <fct> A1, A1, A1, A1, A1, A1, A2, A2, A2, A2, A2, A2
$ Plot <fct> P1, P1, P1, P2, P2, P2, P3, P3, P3, P4, P4, P4
$ Cond <fct> H, M, L, H, M, L, H, M, L, H, M, L
$ Time <int> 1, 2, 3, 4, 1, 2, 3, 4, 1, 2, 3, 4
$ Temp <dbl> 15.73546, 23.83643, 13.64371, 37.95281, 25.29508, 13.79532, 26.87429,...
$ LAT <dbl> 17.25752, 14.07060, 20.74986, 18.41013, 18.46762, 20.38767, 20.14244,...
$ LONG <dbl> 146.2397, 144.8877, 144.6884, 142.0585, 144.0437, 145.8359, 147.7174,...Sorting data
Sorting data (arrange)
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877Sorting by LAT
Between Plot Cond Time Temp LAT LONG
1 A1 P1 M 2 23.83643 14.07060 144.8877
2 A2 P4 L 4 25.89843 14.52130 144.1700
3 A1 P1 H 1 15.73546 17.25752 146.2397
4 A1 P2 H 4 37.95281 18.41013 142.0585
5 A1 P2 M 1 25.29508 18.46762 144.0437
6 A2 P4 M 3 37.11781 18.64913 142.2459
7 A2 P3 M 4 29.38325 19.68780 144.7944
8 A2 P4 H 2 18.94612 20.06427 144.8924
9 A2 P3 H 3 26.87429 20.14244 147.7174
10 A2 P3 L 1 27.75781 20.33795 145.7753
11 A1 P2 L 2 13.79532 20.38767 145.8359
12 A1 P1 L 3 13.64371 20.74986 144.6884Sorting data (arrange)
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877Sorting by LAT (descending order)
Between Plot Cond Time Temp LAT LONG
1 A1 P1 L 3 13.64371 20.74986 144.6884
2 A1 P2 L 2 13.79532 20.38767 145.8359
3 A2 P3 L 1 27.75781 20.33795 145.7753
4 A2 P3 H 3 26.87429 20.14244 147.7174
5 A2 P4 H 2 18.94612 20.06427 144.8924
6 A2 P3 M 4 29.38325 19.68780 144.7944
7 A2 P4 M 3 37.11781 18.64913 142.2459
8 A1 P2 M 1 25.29508 18.46762 144.0437
9 A1 P2 H 4 37.95281 18.41013 142.0585
10 A1 P1 H 1 15.73546 17.25752 146.2397
11 A2 P4 L 4 25.89843 14.52130 144.1700
12 A1 P1 M 2 23.83643 14.07060 144.8877Sorting data (arrange)
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877Sorting by Cond and then TEMP
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A2 P4 H 2 18.94612 20.06427 144.8924
3 A2 P3 H 3 26.87429 20.14244 147.7174
4 A1 P2 H 4 37.95281 18.41013 142.0585
5 A1 P1 L 3 13.64371 20.74986 144.6884
6 A1 P2 L 2 13.79532 20.38767 145.8359
7 A2 P4 L 4 25.89843 14.52130 144.1700
8 A2 P3 L 1 27.75781 20.33795 145.7753
9 A1 P1 M 2 23.83643 14.07060 144.8877
10 A1 P2 M 1 25.29508 18.46762 144.0437
11 A2 P3 M 4 29.38325 19.68780 144.7944
12 A2 P4 M 3 37.11781 18.64913 142.2459Sorting data (arrange)
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877Sort by the sum of Temp and LAT
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P2 L 2 13.79532 20.38767 145.8359
3 A1 P1 L 3 13.64371 20.74986 144.6884
4 A1 P1 M 2 23.83643 14.07060 144.8877
5 A2 P4 H 2 18.94612 20.06427 144.8924
6 A2 P4 L 4 25.89843 14.52130 144.1700
7 A1 P2 M 1 25.29508 18.46762 144.0437
8 A2 P3 H 3 26.87429 20.14244 147.7174
9 A2 P3 L 1 27.75781 20.33795 145.7753
10 A2 P3 M 4 29.38325 19.68780 144.7944
11 A2 P4 M 3 37.11781 18.64913 142.2459
12 A1 P2 H 4 37.95281 18.41013 142.0585Your turn
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877- sort by Between and then Cond
Your turn
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877- sort by Between and then Cond
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P2 H 4 37.95281 18.41013 142.0585
3 A1 P1 L 3 13.64371 20.74986 144.6884
4 A1 P2 L 2 13.79532 20.38767 145.8359
5 A1 P1 M 2 23.83643 14.07060 144.8877
6 A1 P2 M 1 25.29508 18.46762 144.0437
7 A2 P3 H 3 26.87429 20.14244 147.7174
8 A2 P4 H 2 18.94612 20.06427 144.8924
9 A2 P3 L 1 27.75781 20.33795 145.7753
10 A2 P4 L 4 25.89843 14.52130 144.1700
11 A2 P3 M 4 29.38325 19.68780 144.7944
12 A2 P4 M 3 37.11781 18.64913 142.2459Your turn
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877- sort by Condition and then the ratio of Temp to LAT
Your turn
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877- sort by Condition and then the ratio of Temp to LAT
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A2 P4 H 2 18.94612 20.06427 144.8924
3 A2 P3 H 3 26.87429 20.14244 147.7174
4 A1 P2 H 4 37.95281 18.41013 142.0585
5 A1 P1 L 3 13.64371 20.74986 144.6884
6 A1 P2 L 2 13.79532 20.38767 145.8359
7 A2 P3 L 1 27.75781 20.33795 145.7753
8 A2 P4 L 4 25.89843 14.52130 144.1700
9 A1 P2 M 1 25.29508 18.46762 144.0437
10 A2 P3 M 4 29.38325 19.68780 144.7944
11 A1 P1 M 2 23.83643 14.07060 144.8877
12 A2 P4 M 3 37.11781 18.64913 142.2459Manipulating factors
Manipulating factors
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877[1] "H" "L" "M"- re-levelling
- re-labelling
- technically these operations are performed on single variables (vectors)
Re-levelling (sorting) factors
[1] "H" "L" "M"[1] "L" "M" "H" Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877
3 A1 P1 L 3 13.64371 20.74986 144.6884
4 A1 P2 H 4 37.95281 18.41013 142.0585
5 A1 P2 M 1 25.29508 18.46762 144.0437
6 A1 P2 L 2 13.79532 20.38767 145.8359Re-levelling (sorting) factors
[1] "H" "L" "M"[1] "High" "Low" "Medium" Between Plot Cond Time Temp LAT LONG
1 A1 P1 High 1 15.73546 17.25752 146.2397
2 A1 P1 Medium 2 23.83643 14.07060 144.8877
3 A1 P1 Low 3 13.64371 20.74986 144.6884
4 A1 P2 High 4 37.95281 18.41013 142.0585
5 A1 P2 Medium 1 25.29508 18.46762 144.0437
6 A1 P2 Low 2 13.79532 20.38767 145.8359Re-levelling (sorting) factors
[1] "H" "L" "M"> data.3$Cond <- factor(data.3$Cond, levels=c('L','M','H'),
+ labels=c("Low","Medium","High"))
> levels(data.3$Cond)[1] "Low" "Medium" "High" Between Plot Cond Time Temp LAT LONG
1 A1 P1 High 1 15.73546 17.25752 146.2397
2 A1 P1 Medium 2 23.83643 14.07060 144.8877
3 A1 P1 Low 3 13.64371 20.74986 144.6884
4 A1 P2 High 4 37.95281 18.41013 142.0585
5 A1 P2 Medium 1 25.29508 18.46762 144.0437
6 A1 P2 Low 2 13.79532 20.38767 145.8359Re-labelling factors
[1] "H" "Low" "Medium" Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 Medium 2 23.83643 14.07060 144.8877
3 A1 P1 Low 3 13.64371 20.74986 144.6884
4 A1 P2 H 4 37.95281 18.41013 142.0585
5 A1 P2 Medium 1 25.29508 18.46762 144.0437
6 A1 P2 Low 2 13.79532 20.38767 145.8359
7 A2 P3 H 3 26.87429 20.14244 147.7174
8 A2 P3 Medium 4 29.38325 19.68780 144.7944
9 A2 P3 Low 1 27.75781 20.33795 145.7753
10 A2 P4 H 2 18.94612 20.06427 144.8924
11 A2 P4 Medium 3 37.11781 18.64913 142.2459
12 A2 P4 Low 4 25.89843 14.52130 144.1700Re-levelling & labelling
> data.3 <- data.1 %>% mutate(Cond=recode_factor(Cond,'L'='Low', 'M'='Medium'))
> levels(data.3$Cond) [1] "Low" "Medium" "H" Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 Medium 2 23.83643 14.07060 144.8877
3 A1 P1 Low 3 13.64371 20.74986 144.6884
4 A1 P2 H 4 37.95281 18.41013 142.0585
5 A1 P2 Medium 1 25.29508 18.46762 144.0437
6 A1 P2 Low 2 13.79532 20.38767 145.8359
7 A2 P3 H 3 26.87429 20.14244 147.7174
8 A2 P3 Medium 4 29.38325 19.68780 144.7944
9 A2 P3 Low 1 27.75781 20.33795 145.7753
10 A2 P4 H 2 18.94612 20.06427 144.8924
11 A2 P4 Medium 3 37.11781 18.64913 142.2459
12 A2 P4 Low 4 25.89843 14.52130 144.1700Re-levelling & labelling
You might also want to check out the forcats package
Subset columns
Selecting columns (select)
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877 Between Plot Cond Time Temp
1 A1 P1 H 1 15.73546
2 A1 P1 M 2 23.83643
3 A1 P1 L 3 13.64371
4 A1 P2 H 4 37.95281
5 A1 P2 M 1 25.29508
6 A1 P2 L 2 13.79532
7 A2 P3 H 3 26.87429
8 A2 P3 M 4 29.38325
9 A2 P3 L 1 27.75781
10 A2 P4 H 2 18.94612
11 A2 P4 M 3 37.11781
12 A2 P4 L 4 25.89843Selecting columns (select)
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877 Between Plot Cond Time Temp
1 A1 P1 H 1 15.73546
2 A1 P1 M 2 23.83643
3 A1 P1 L 3 13.64371
4 A1 P2 H 4 37.95281
5 A1 P2 M 1 25.29508
6 A1 P2 L 2 13.79532
7 A2 P3 H 3 26.87429
8 A2 P3 M 4 29.38325
9 A2 P3 L 1 27.75781
10 A2 P4 H 2 18.94612
11 A2 P4 M 3 37.11781
12 A2 P4 L 4 25.89843Selecting columns (select)
helper functions
contains()ends_with()starts_with()matches()everything()- …
must evaluate to indices
Selecting columns (select)
helper functions
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877 Plot LAT LONG
1 P1 17.25752 146.2397
2 P1 14.07060 144.8877
3 P1 20.74986 144.6884
4 P2 18.41013 142.0585
5 P2 18.46762 144.0437
6 P2 20.38767 145.8359
7 P3 20.14244 147.7174
8 P3 19.68780 144.7944
9 P3 20.33795 145.7753
10 P4 20.06427 144.8924
11 P4 18.64913 142.2459
12 P4 14.52130 144.1700Selecting columns (select)
helper functions
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877 LAT LONG
1 17.25752 146.2397
2 14.07060 144.8877
3 20.74986 144.6884
4 18.41013 142.0585
5 18.46762 144.0437
6 20.38767 145.8359
7 20.14244 147.7174
8 19.68780 144.7944
9 20.33795 145.7753
10 20.06427 144.8924
11 18.64913 142.2459
12 14.52130 144.1700Selecting columns (select)
helper functions
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877 Plot LAT
1 P1 17.25752
2 P1 14.07060
3 P1 20.74986
4 P2 18.41013
5 P2 18.46762
6 P2 20.38767
7 P3 20.14244
8 P3 19.68780
9 P3 20.33795
10 P4 20.06427
11 P4 18.64913
12 P4 14.52130Selecting columns (select)
helper functions
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877 Time Temp
1 1 15.73546
2 2 23.83643
3 3 13.64371
4 4 37.95281
5 1 25.29508
6 2 13.79532
7 3 26.87429
8 4 29.38325
9 1 27.75781
10 2 18.94612
11 3 37.11781
12 4 25.89843Regular expressions (regexp)
https://www.rstudio.com/resources/cheatsheets/raw/master/regex.pdf
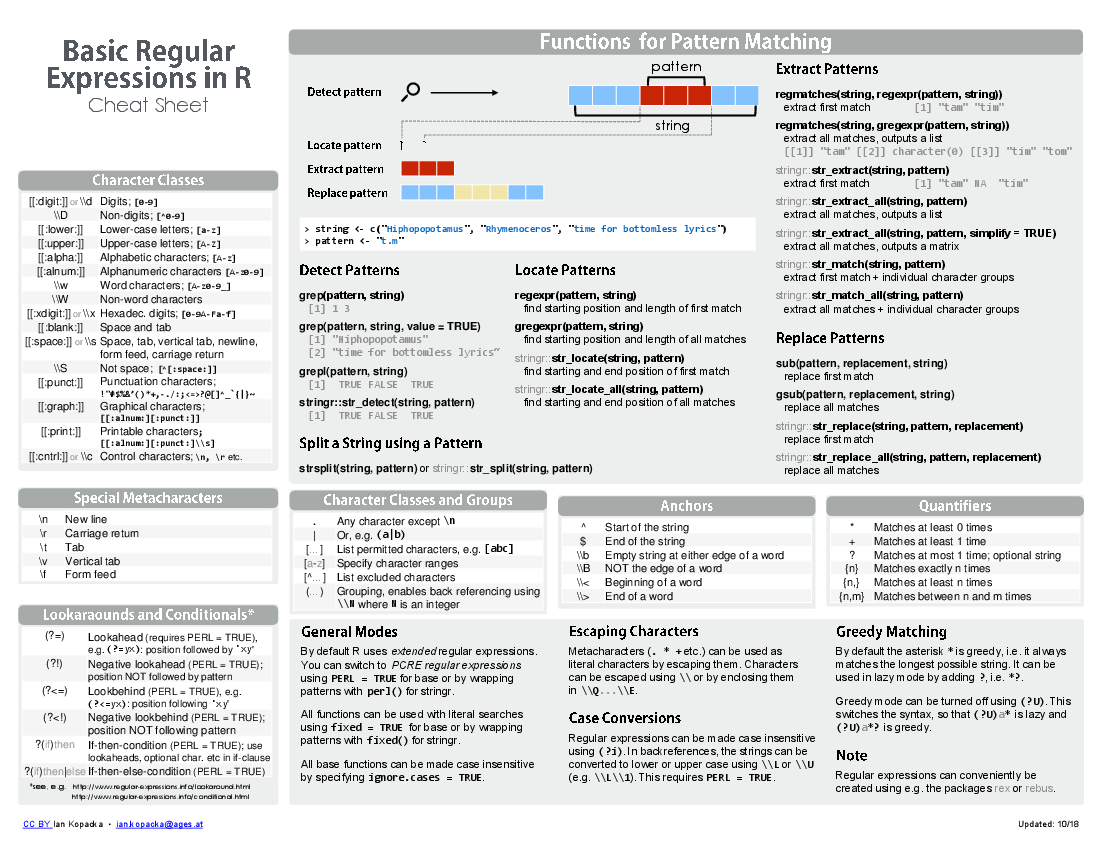
<img src=“figure/regex.png”, width=“550px”>
Selecting columns (select)
helper functions
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877 Between Plot Cond Time Temp
1 A1 P1 H 1 15.73546
2 A1 P1 M 2 23.83643
3 A1 P1 L 3 13.64371
4 A1 P2 H 4 37.95281
5 A1 P2 M 1 25.29508
6 A1 P2 L 2 13.79532
7 A2 P3 H 3 26.87429
8 A2 P3 M 4 29.38325
9 A2 P3 L 1 27.75781
10 A2 P4 H 2 18.94612
11 A2 P4 M 3 37.11781
12 A2 P4 L 4 25.89843Your turn
lat long month year cloudhigh cloudlow
1 36.20000 -113.8 1 1995 26.0 7.5
2 33.70435 -113.8 1 1995 20.0 11.5
3 31.20870 -113.8 1 1995 16.0 16.5
4 28.71304 -113.8 1 1995 13.0 20.5
5 26.21739 -113.8 1 1995 7.5 26.0
6 23.72174 -113.8 1 1995 8.0 30.0
cloudmid ozone pressure surftemp temperature
1 34.5 304 835 272.7 272.1
2 32.5 304 940 279.5 282.2
3 26.0 298 960 284.7 285.2
4 14.5 276 990 289.3 290.7
5 10.5 274 1000 292.2 292.7
6 9.5 264 1000 294.1 293.6Select lat, long, and cloud.. columns
Your turn
lat long month year cloudhigh cloudlow cloudmid ozone pressure surftemp temperature
1 36.20000 -113.8 1 1995 26.0 7.5 34.5 304 835 272.7 272.1
2 33.70435 -113.8 1 1995 20.0 11.5 32.5 304 940 279.5 282.2
3 31.20870 -113.8 1 1995 16.0 16.5 26.0 298 960 284.7 285.2
4 28.71304 -113.8 1 1995 13.0 20.5 14.5 276 990 289.3 290.7
5 26.21739 -113.8 1 1995 7.5 26.0 10.5 274 1000 292.2 292.7
6 23.72174 -113.8 1 1995 8.0 30.0 9.5 264 1000 294.1 293.6 lat long cloudhigh cloudlow cloudmid
1 36.20000 -113.8 26.0 7.5 34.5
2 33.70435 -113.8 20.0 11.5 32.5
3 31.20870 -113.8 16.0 16.5 26.0
4 28.71304 -113.8 13.0 20.5 14.5
5 26.21739 -113.8 7.5 26.0 10.5
6 23.72174 -113.8 8.0 30.0 9.5Your turn
Psammocora contigua Psammocora digitata Pocillopora damicornis time rep
V1 0 0 79 81 1
V2 0 0 51 81 2
V3 0 0 42 81 3
V4 0 0 15 81 4
V5 0 0 9 81 5
V6 0 0 72 81 6
V7 0 0 0 81 7
V8 0 0 16 81 8
V9 0 0 0 81 9
V10 0 0 16 81 10Select rep, time and only Species that DONT contain pora
Your turn
Select rep, time and only Species that DONT contain pora
Select awkward names
Pocillopora damicornis
V1 79
V2 51
V3 42
V4 15
V5 9
V6 72
V7 0
V8 16
V9 0
V10 16
V11 0
V12 0
V13 0
V14 0
V15 0
V16 0
V17 0
V18 0
V19 0
V20 0
V21 0
V22 0
V23 0
V24 0
V25 0
V26 0
V27 0
V28 0
V29 0
V30 0
V31 0
V32 0
V33 0
V34 0
V35 0
V36 0
V37 0
V38 0
V39 0
V40 0
V41 18
V42 0
V43 0
V44 0
V45 0
V46 0
V47 0
V48 0
V49 0
V50 0
V51 0
V52 0
V53 0
V54 0
V55 0
V56 0
V57 10
V58 0
V59 30
V60 0Re-naming columns (vectors)
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877 Between Plot Condition Time Temperature LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877
3 A1 P1 L 3 13.64371 20.74986 144.6884
4 A1 P2 H 4 37.95281 18.41013 142.0585
5 A1 P2 M 1 25.29508 18.46762 144.0437
6 A1 P2 L 2 13.79532 20.38767 145.8359
7 A2 P3 H 3 26.87429 20.14244 147.7174
8 A2 P3 M 4 29.38325 19.68780 144.7944
9 A2 P3 L 1 27.75781 20.33795 145.7753
10 A2 P4 H 2 18.94612 20.06427 144.8924
11 A2 P4 M 3 37.11781 18.64913 142.2459
12 A2 P4 L 4 25.89843 14.52130 144.1700Filtering
Filtering
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877 Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P2 H 4 37.95281 18.41013 142.0585
3 A2 P3 H 3 26.87429 20.14244 147.7174
4 A2 P4 H 2 18.94612 20.06427 144.8924 Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877
3 A1 P2 H 4 37.95281 18.41013 142.0585
4 A1 P2 M 1 25.29508 18.46762 144.0437
5 A2 P3 H 3 26.87429 20.14244 147.7174
6 A2 P3 M 4 29.38325 19.68780 144.7944
7 A2 P4 H 2 18.94612 20.06427 144.8924
8 A2 P4 M 3 37.11781 18.64913 142.2459Filtering
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877 Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A2 P4 H 2 18.94612 20.06427 144.8924 Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877
3 A1 P1 L 3 13.64371 20.74986 144.6884
4 A1 P2 H 4 37.95281 18.41013 142.0585
5 A1 P2 L 2 13.79532 20.38767 145.8359
6 A2 P3 H 3 26.87429 20.14244 147.7174
7 A2 P4 H 2 18.94612 20.06427 144.8924Your turn
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877Keep only those rows with Temp less than 20 and LAT greater than 20 or LONG less than 145
Your turn
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877Keep only those rows with Temp less than 20 and LAT greater than 20, or LONG less than 145
Between Plot Cond Time Temp LAT LONG
1 A1 P1 L 3 13.64371 20.74986 144.6884
2 A1 P2 L 2 13.79532 20.38767 145.8359
3 A2 P4 H 2 18.94612 20.06427 144.8924Your turn
Observations: 41,472
Variables: 11
$ lat <dbl> 36.200000, 33.704348, 31.208696, 28.713043, 26.217391, 23.721739,...
$ long <dbl> -113.8000, -113.8000, -113.8000, -113.8000, -113.8000, -113.8000,...
$ month <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,...
$ year <int> 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,...
$ cloudhigh <dbl> 26.0, 20.0, 16.0, 13.0, 7.5, 8.0, 14.5, 19.5, 22.5, 21.0, 19.0, 1...
$ cloudlow <dbl> 7.5, 11.5, 16.5, 20.5, 26.0, 30.0, 29.5, 26.5, 27.5, 26.0, 28.5, ...
$ cloudmid <dbl> 34.5, 32.5, 26.0, 14.5, 10.5, 9.5, 11.0, 17.5, 18.5, 16.5, 12.5, ...
$ ozone <dbl> 304, 304, 298, 276, 274, 264, 258, 252, 250, 250, 248, 248, 250, ...
$ pressure <dbl> 835, 940, 960, 990, 1000, 1000, 1000, 1000, 1000, 1000, 1000, 100...
$ surftemp <dbl> 272.7, 279.5, 284.7, 289.3, 292.2, 294.1, 295.0, 298.3, 300.1, 30...
$ temperature <dbl> 272.1, 282.2, 285.2, 290.7, 292.7, 293.6, 294.6, 296.9, 297.8, 29...Filter to the largest ozone value for the second month of the last year
Your turn
Filter to the largest ozone value for the second month of the last year
Your turn
Filter to all ozone values between 320 and 325 in the first month of the last year
Observations: 41,472
Variables: 11
$ lat <dbl> 36.200000, 33.704348, 31.208696, 28.713043, 26.217391, 23.721739,...
$ long <dbl> -113.8000, -113.8000, -113.8000, -113.8000, -113.8000, -113.8000,...
$ month <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,...
$ year <int> 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,...
$ cloudhigh <dbl> 26.0, 20.0, 16.0, 13.0, 7.5, 8.0, 14.5, 19.5, 22.5, 21.0, 19.0, 1...
$ cloudlow <dbl> 7.5, 11.5, 16.5, 20.5, 26.0, 30.0, 29.5, 26.5, 27.5, 26.0, 28.5, ...
$ cloudmid <dbl> 34.5, 32.5, 26.0, 14.5, 10.5, 9.5, 11.0, 17.5, 18.5, 16.5, 12.5, ...
$ ozone <dbl> 304, 304, 298, 276, 274, 264, 258, 252, 250, 250, 248, 248, 250, ...
$ pressure <dbl> 835, 940, 960, 990, 1000, 1000, 1000, 1000, 1000, 1000, 1000, 100...
$ surftemp <dbl> 272.7, 279.5, 284.7, 289.3, 292.2, 294.1, 295.0, 298.3, 300.1, 30...
$ temperature <dbl> 272.1, 282.2, 285.2, 290.7, 292.7, 293.6, 294.6, 296.9, 297.8, 29...Your turn
Filter to all ozone values between 320 and 325 in the first month of the last year
Slicing
Filtering by row number
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877 Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877
3 A1 P1 L 3 13.64371 20.74986 144.6884
4 A1 P2 H 4 37.95281 18.41013 142.0585 Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877
3 A1 P1 L 3 13.64371 20.74986 144.6884
4 A1 P2 H 4 37.95281 18.41013 142.0585
5 A2 P3 H 3 26.87429 20.14244 147.7174Sampling
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877 Between Plot Cond Time Temp LAT LONG
5 A1 P2 M 1 25.29508 18.46762 144.0437
5.1 A1 P2 M 1 25.29508 18.46762 144.0437
6 A1 P2 L 2 13.79532 20.38767 145.8359
11 A2 P4 M 3 37.11781 18.64913 142.2459
11.1 A2 P4 M 3 37.11781 18.64913 142.2459
5.2 A1 P2 M 1 25.29508 18.46762 144.0437
10 A2 P4 H 2 18.94612 20.06427 144.8924
12 A2 P4 L 4 25.89843 14.52130 144.1700
6.1 A1 P2 L 2 13.79532 20.38767 145.8359
9 A2 P3 L 1 27.75781 20.33795 145.7753Sampling
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877 Between Plot Cond Time Temp LAT LONG
5 A1 P2 M 1 25.29508 18.46762 144.0437
4 A1 P2 H 4 37.95281 18.41013 142.0585
10 A2 P4 H 2 18.94612 20.06427 144.8924
3 A1 P1 L 3 13.64371 20.74986 144.6884
9 A2 P3 L 1 27.75781 20.33795 145.7753
2 A1 P1 M 2 23.83643 14.07060 144.8877Effects of filtering
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877[1] "H" "L" "M"Effects of filtering
> #subset the dataset to just Cond H
> data.3<-filter(data.1,Plot=='P1')
> #examine subset data
> data.3 Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877
3 A1 P1 L 3 13.64371 20.74986 144.6884[1] "H" "L" "M"[1] "P1" "P2" "P3" "P4"[1] "A1" "A2"Effects of filtering
Correction - all factors
> #subset the dataset to just Cond H
> data.3<-filter(data.1,Plot=='P1')
> #drop the unused factor levels from all factors
> data.3<-droplevels(data.3)
> #examine the levels of each factor
> levels(data.3$Cond)[1] "H" "L" "M"[1] "P1"[1] "A1"Effects of filtering
Correction - single factor
> #subset the dataset to just Cond H
> data.3<-filter(data.1,Plot=='P1')
> #drop the unused factor levels from Cond
> data.3$Plot<-factor(data.3$Plot)
> #examine the levels of each factor
> levels(data.3$Cond)[1] "H" "L" "M"[1] "P1"[1] "A1" "A2"Adding columns
Mutate
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877 Between Plot Cond Time Temp LAT LONG LL
1 A1 P1 H 1 15.73546 17.25752 146.2397 163.4972
2 A1 P1 M 2 23.83643 14.07060 144.8877 158.9583
3 A1 P1 L 3 13.64371 20.74986 144.6884 165.4383
4 A1 P2 H 4 37.95281 18.41013 142.0585 160.4686
5 A1 P2 M 1 25.29508 18.46762 144.0437 162.5113
6 A1 P2 L 2 13.79532 20.38767 145.8359 166.2236
7 A2 P3 H 3 26.87429 20.14244 147.7174 167.8598
8 A2 P3 M 4 29.38325 19.68780 144.7944 164.4822
9 A2 P3 L 1 27.75781 20.33795 145.7753 166.1133
10 A2 P4 H 2 18.94612 20.06427 144.8924 164.9567
11 A2 P4 M 3 37.11781 18.64913 142.2459 160.8950
12 A2 P4 L 4 25.89843 14.52130 144.1700 158.6913Mutate
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877 Between Plot Cond Time Temp LAT LONG logTemp
1 A1 P1 H 1 15.73546 17.25752 146.2397 2.755917
2 A1 P1 M 2 23.83643 14.07060 144.8877 3.171215
3 A1 P1 L 3 13.64371 20.74986 144.6884 2.613279
4 A1 P2 H 4 37.95281 18.41013 142.0585 3.636343
5 A1 P2 M 1 25.29508 18.46762 144.0437 3.230610
6 A1 P2 L 2 13.79532 20.38767 145.8359 2.624329
7 A2 P3 H 3 26.87429 20.14244 147.7174 3.291170
8 A2 P3 M 4 29.38325 19.68780 144.7944 3.380425
9 A2 P3 L 1 27.75781 20.33795 145.7753 3.323517
10 A2 P4 H 2 18.94612 20.06427 144.8924 2.941599
11 A2 P4 M 3 37.11781 18.64913 142.2459 3.614097
12 A2 P4 L 4 25.89843 14.52130 144.1700 3.254182Mutate
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877> mutate(data.1, MeanTemp=mean(Temp), cTemp=Temp-MeanTemp)
> ## OR if just want the centered variable..
> #mutate(data.1, cTemp=Temp-mean(Temp)) Between Plot Cond Time Temp LAT LONG MeanTemp cTemp
1 A1 P1 H 1 15.73546 17.25752 146.2397 24.68638 -8.9509150
2 A1 P1 M 2 23.83643 14.07060 144.8877 24.68638 -0.8499436
3 A1 P1 L 3 13.64371 20.74986 144.6884 24.68638 -11.0426630
4 A1 P2 H 4 37.95281 18.41013 142.0585 24.68638 13.2664312
5 A1 P2 M 1 25.29508 18.46762 144.0437 24.68638 0.6087009
6 A1 P2 L 2 13.79532 20.38767 145.8359 24.68638 -10.8910607
7 A2 P3 H 3 26.87429 20.14244 147.7174 24.68638 2.1879137
8 A2 P3 M 4 29.38325 19.68780 144.7944 24.68638 4.6968702
9 A2 P3 L 1 27.75781 20.33795 145.7753 24.68638 3.0714367
10 A2 P4 H 2 18.94612 20.06427 144.8924 24.68638 -5.7402607
11 A2 P4 M 3 37.11781 18.64913 142.2459 24.68638 12.4314348
12 A2 P4 L 4 25.89843 14.52130 144.1700 24.68638 1.2120555Mutate
Window functions
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877 Between Plot Cond Time Temp LAT LONG leadTemp lagTemp
1 A1 P1 H 1 15.73546 17.25752 146.2397 23.83643 NA
2 A1 P1 M 2 23.83643 14.07060 144.8877 13.64371 15.73546
3 A1 P1 L 3 13.64371 20.74986 144.6884 37.95281 23.83643
4 A1 P2 H 4 37.95281 18.41013 142.0585 25.29508 13.64371
5 A1 P2 M 1 25.29508 18.46762 144.0437 13.79532 37.95281
6 A1 P2 L 2 13.79532 20.38767 145.8359 26.87429 25.29508
7 A2 P3 H 3 26.87429 20.14244 147.7174 29.38325 13.79532
8 A2 P3 M 4 29.38325 19.68780 144.7944 27.75781 26.87429
9 A2 P3 L 1 27.75781 20.33795 145.7753 18.94612 29.38325
10 A2 P4 H 2 18.94612 20.06427 144.8924 37.11781 27.75781
11 A2 P4 M 3 37.11781 18.64913 142.2459 25.89843 18.94612
12 A2 P4 L 4 25.89843 14.52130 144.1700 NA 37.11781Mutate
Window functions
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877 Between Plot Cond Time Temp LAT LONG rankTime denseRankTime
1 A1 P1 H 1 15.73546 17.25752 146.2397 1 1
2 A1 P1 M 2 23.83643 14.07060 144.8877 4 2
3 A1 P1 L 3 13.64371 20.74986 144.6884 7 3
4 A1 P2 H 4 37.95281 18.41013 142.0585 10 4
5 A1 P2 M 1 25.29508 18.46762 144.0437 1 1
6 A1 P2 L 2 13.79532 20.38767 145.8359 4 2
7 A2 P3 H 3 26.87429 20.14244 147.7174 7 3
8 A2 P3 M 4 29.38325 19.68780 144.7944 10 4
9 A2 P3 L 1 27.75781 20.33795 145.7753 1 1
10 A2 P4 H 2 18.94612 20.06427 144.8924 4 2
11 A2 P4 M 3 37.11781 18.64913 142.2459 7 3
12 A2 P4 L 4 25.89843 14.52130 144.1700 10 4Mutate
Window functions
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877 Between Plot Cond Time Temp LAT LONG rowTemp rowTime
1 A1 P1 H 1 15.73546 17.25752 146.2397 3 1
2 A1 P1 M 2 23.83643 14.07060 144.8877 5 4
3 A1 P1 L 3 13.64371 20.74986 144.6884 1 7
4 A1 P2 H 4 37.95281 18.41013 142.0585 12 10
5 A1 P2 M 1 25.29508 18.46762 144.0437 6 2
6 A1 P2 L 2 13.79532 20.38767 145.8359 2 5
7 A2 P3 H 3 26.87429 20.14244 147.7174 8 8
8 A2 P3 M 4 29.38325 19.68780 144.7944 10 11
9 A2 P3 L 1 27.75781 20.33795 145.7753 9 3
10 A2 P4 H 2 18.94612 20.06427 144.8924 4 6
11 A2 P4 M 3 37.11781 18.64913 142.2459 11 9
12 A2 P4 L 4 25.89843 14.52130 144.1700 7 12Mutate
Window functions
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877 Between Plot Cond Time Temp LAT LONG ntile(Temp, 4)
1 A1 P1 H 1 15.73546 17.25752 146.2397 1
2 A1 P1 M 2 23.83643 14.07060 144.8877 2
3 A1 P1 L 3 13.64371 20.74986 144.6884 1
4 A1 P2 H 4 37.95281 18.41013 142.0585 4
5 A1 P2 M 1 25.29508 18.46762 144.0437 2
6 A1 P2 L 2 13.79532 20.38767 145.8359 1
7 A2 P3 H 3 26.87429 20.14244 147.7174 3
8 A2 P3 M 4 29.38325 19.68780 144.7944 4
9 A2 P3 L 1 27.75781 20.33795 145.7753 3
10 A2 P4 H 2 18.94612 20.06427 144.8924 2
11 A2 P4 M 3 37.11781 18.64913 142.2459 4
12 A2 P4 L 4 25.89843 14.52130 144.1700 3Mutate
Window functions
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877 Between Plot Cond Time Temp LAT LONG between(Temp, 20, 30)
1 A1 P1 H 1 15.73546 17.25752 146.2397 FALSE
2 A1 P1 M 2 23.83643 14.07060 144.8877 TRUE
3 A1 P1 L 3 13.64371 20.74986 144.6884 FALSE
4 A1 P2 H 4 37.95281 18.41013 142.0585 FALSE
5 A1 P2 M 1 25.29508 18.46762 144.0437 TRUE
6 A1 P2 L 2 13.79532 20.38767 145.8359 FALSE
7 A2 P3 H 3 26.87429 20.14244 147.7174 TRUE
8 A2 P3 M 4 29.38325 19.68780 144.7944 TRUE
9 A2 P3 L 1 27.75781 20.33795 145.7753 TRUE
10 A2 P4 H 2 18.94612 20.06427 144.8924 FALSE
11 A2 P4 M 3 37.11781 18.64913 142.2459 FALSE
12 A2 P4 L 4 25.89843 14.52130 144.1700 TRUEMutate
Window functions
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877> mutate(data.1, fTemp=ifelse(Temp<20, 'Low',
+ ifelse(between(Temp,20,30), 'Medium', 'High')))
> ## OR
> mutate(data.1, fTemp=case_when(Temp<20 ~ 'Low',
+ between(Temp, 20, 30) ~ 'Medium',
+ Temp>30 ~ 'High')) Between Plot Cond Time Temp LAT LONG fTemp
1 A1 P1 H 1 15.73546 17.25752 146.2397 Low
2 A1 P1 M 2 23.83643 14.07060 144.8877 Medium
3 A1 P1 L 3 13.64371 20.74986 144.6884 Low
4 A1 P2 H 4 37.95281 18.41013 142.0585 High
5 A1 P2 M 1 25.29508 18.46762 144.0437 Medium
6 A1 P2 L 2 13.79532 20.38767 145.8359 Low
7 A2 P3 H 3 26.87429 20.14244 147.7174 Medium
8 A2 P3 M 4 29.38325 19.68780 144.7944 Medium
9 A2 P3 L 1 27.75781 20.33795 145.7753 Medium
10 A2 P4 H 2 18.94612 20.06427 144.8924 Low
11 A2 P4 M 3 37.11781 18.64913 142.2459 High
12 A2 P4 L 4 25.89843 14.52130 144.1700 Medium
Between Plot Cond Time Temp LAT LONG fTemp
1 A1 P1 H 1 15.73546 17.25752 146.2397 Low
2 A1 P1 M 2 23.83643 14.07060 144.8877 Medium
3 A1 P1 L 3 13.64371 20.74986 144.6884 Low
4 A1 P2 H 4 37.95281 18.41013 142.0585 High
5 A1 P2 M 1 25.29508 18.46762 144.0437 Medium
6 A1 P2 L 2 13.79532 20.38767 145.8359 Low
7 A2 P3 H 3 26.87429 20.14244 147.7174 Medium
8 A2 P3 M 4 29.38325 19.68780 144.7944 Medium
9 A2 P3 L 1 27.75781 20.33795 145.7753 Medium
10 A2 P4 H 2 18.94612 20.06427 144.8924 Low
11 A2 P4 M 3 37.11781 18.64913 142.2459 High
12 A2 P4 L 4 25.89843 14.52130 144.1700 MediumMutate
Window functions
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877 Between Plot Cond Time Temp LAT LONG fTemp
1 A1 P1 H 1 15.73546 17.25752 146.2397 Low
2 A1 P1 M 2 23.83643 14.07060 144.8877 Medium
3 A1 P1 L 3 13.64371 20.74986 144.6884 Low
4 A1 P2 H 4 37.95281 18.41013 142.0585 High
5 A1 P2 M 1 25.29508 18.46762 144.0437 Medium
6 A1 P2 L 2 13.79532 20.38767 145.8359 Low
7 A2 P3 H 3 26.87429 20.14244 147.7174 Medium
8 A2 P3 M 4 29.38325 19.68780 144.7944 Medium
9 A2 P3 L 1 27.75781 20.33795 145.7753 Medium
10 A2 P4 H 2 18.94612 20.06427 144.8924 Low
11 A2 P4 M 3 37.11781 18.64913 142.2459 High
12 A2 P4 L 4 25.89843 14.52130 144.1700 MediumSummarising (aggregating) data
Summarise
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877 MeanTemp VarTemp N
1 24.68638 65.72792 12> SE <- function(x) sd(x)/sqrt(length(x))
> summarise(data.1, MeanTemp=mean(Temp), VarTemp=var(Temp), SEM=SE(Temp)) MeanTemp VarTemp SEM
1 24.68638 65.72792 2.340369Summarise
Between_mean Plot_mean Cond_mean Time_mean Temp_mean LAT_mean LONG_mean Between_var
1 NA NA NA 2.5 24.68638 18.56219 144.7791 0.2727273
Plot_var Cond_var Time_var Temp_var LAT_var LONG_var
1 1.363636 0.7272727 1.363636 65.72792 5.048825 2.512696 Temp_mean LAT_mean Temp_var LAT_var
1 24.68638 18.56219 65.72792 5.048825 Time_mean Temp_mean LAT_mean LONG_mean Time_var Temp_var LAT_var LONG_var
1 2.5 24.68638 18.56219 144.7791 1.363636 65.72792 5.048825 2.512696Summarise
# A tibble: 3 x 2
Cond n
<fct> <int>
1 H 4
2 L 4
3 M 4# A tibble: 6 x 3
Cond `between(Temp, 20, 30)` n
<fct> <lgl> <int>
1 H FALSE 3
2 H TRUE 1
3 L FALSE 2
4 L TRUE 2
5 M FALSE 1
6 M TRUE 3Piping
Piping
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877
3 A1 P1 L 3 13.64371 20.74986 144.6884
4 A1 P2 H 4 37.95281 18.41013 142.0585
5 A1 P2 M 1 25.29508 18.46762 144.0437
6 A1 P2 L 2 13.79532 20.38767 145.8359 Cond Time Temp
1 H 1 15.73546
2 H 4 37.95281
3 H 3 26.87429
4 H 2 18.94612Grouping (=aggregating)
Grouping
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877
3 A1 P1 L 3 13.64371 20.74986 144.6884
4 A1 P2 H 4 37.95281 18.41013 142.0585
5 A1 P2 M 1 25.29508 18.46762 144.0437
6 A1 P2 L 2 13.79532 20.38767 145.8359# A tibble: 4 x 3
# Groups: Between [?]
Between Plot Mean
<fct> <fct> <dbl>
1 A1 P1 17.7
2 A1 P2 25.7
3 A2 P3 28.0
4 A2 P4 27.3Grouping
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877
3 A1 P1 L 3 13.64371 20.74986 144.6884
4 A1 P2 H 4 37.95281 18.41013 142.0585
5 A1 P2 M 1 25.29508 18.46762 144.0437
6 A1 P2 L 2 13.79532 20.38767 145.8359> data.1 %>% group_by(Between,Plot) %>%
+ summarise(Mean=mean(Temp), Var=var(Temp), N=n(),First=first(Temp))# A tibble: 4 x 6
# Groups: Between [?]
Between Plot Mean Var N First
<fct> <fct> <dbl> <dbl> <int> <dbl>
1 A1 P1 17.7 29.0 3 15.7
2 A1 P2 25.7 146. 3 38.0
3 A2 P3 28.0 1.62 3 26.9
4 A2 P4 27.3 84.1 3 18.9Grouping
mutate vs summarise
# A tibble: 4 x 3
# Groups: Between [?]
Between Plot Mean
<fct> <fct> <dbl>
1 A1 P1 17.7
2 A1 P2 25.7
3 A2 P3 28.0
4 A2 P4 27.3# A tibble: 12 x 8
# Groups: Between, Plot [4]
Between Plot Cond Time Temp LAT LONG Mean
<fct> <fct> <fct> <int> <dbl> <dbl> <dbl> <dbl>
1 A1 P1 H 1 15.7 17.3 146. 17.7
2 A1 P1 M 2 23.8 14.1 145. 17.7
3 A1 P1 L 3 13.6 20.7 145. 17.7
4 A1 P2 H 4 38.0 18.4 142. 25.7
5 A1 P2 M 1 25.3 18.5 144. 25.7
6 A1 P2 L 2 13.8 20.4 146. 25.7
7 A2 P3 H 3 26.9 20.1 148. 28.0
8 A2 P3 M 4 29.4 19.7 145. 28.0
9 A2 P3 L 1 27.8 20.3 146. 28.0
10 A2 P4 H 2 18.9 20.1 145. 27.3
11 A2 P4 M 3 37.1 18.6 142. 27.3
12 A2 P4 L 4 25.9 14.5 144. 27.3Grouping
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877# A tibble: 12 x 9
# Groups: Between, Plot [4]
Between Plot Cond Time Temp LAT LONG Mean cTemp
<fct> <fct> <fct> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
1 A1 P1 H 1 15.7 17.3 146. 17.7 -2.00
2 A1 P1 M 2 23.8 14.1 145. 17.7 6.10
3 A1 P1 L 3 13.6 20.7 145. 17.7 -4.09
4 A1 P2 H 4 38.0 18.4 142. 25.7 12.3
5 A1 P2 M 1 25.3 18.5 144. 25.7 -0.386
6 A1 P2 L 2 13.8 20.4 146. 25.7 -11.9
7 A2 P3 H 3 26.9 20.1 148. 28.0 -1.13
8 A2 P3 M 4 29.4 19.7 145. 28.0 1.38
9 A2 P3 L 1 27.8 20.3 146. 28.0 -0.247
10 A2 P4 H 2 18.9 20.1 145. 27.3 -8.37
11 A2 P4 M 3 37.1 18.6 142. 27.3 9.80
12 A2 P4 L 4 25.9 14.5 144. 27.3 -1.42 Grouping
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877# A tibble: 4 x 7
# Groups: Between [?]
Between Plot Cond Time Temp LAT LONG
<fct> <fct> <dbl> <dbl> <dbl> <dbl> <dbl>
1 A1 P1 NA 2.00 17.7 17.4 145.
2 A1 P2 NA 2.33 25.7 19.1 144.
3 A2 P3 NA 2.67 28.0 20.1 146.
4 A2 P4 NA 3.00 27.3 17.7 144.Grouping
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877# A tibble: 4 x 5
# Groups: Between [?]
Between Plot Temp LAT LONG
<fct> <fct> <dbl> <dbl> <dbl>
1 A1 P1 17.7 17.4 145.
2 A1 P2 25.7 19.1 144.
3 A2 P3 28.0 20.1 146.
4 A2 P4 27.3 17.7 144.Grouping
Between Plot Cond Time Temp LAT LONG
1 A1 P1 H 1 15.73546 17.25752 146.2397
2 A1 P1 M 2 23.83643 14.07060 144.8877# A tibble: 4 x 8
# Groups: Between [?]
Between Plot Temp_mean LAT_mean LONG_mean Temp_SE LAT_SE LONG_SE
<fct> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 A1 P1 17.7 17.4 145. 3.11 1.93 0.487
2 A1 P2 25.7 19.1 144. 6.98 0.650 1.09
3 A2 P3 28.0 20.1 146. 0.735 0.193 0.859
4 A2 P4 27.3 17.7 144. 5.29 1.66 0.790Your turn
Calculate for each year, the mean abundance of Pocillopora damicornis
Psammocora contigua Psammocora digitata Pocillopora damicornis time rep
V1 0 0 79 81 1
V2 0 0 51 81 2
V3 0 0 42 81 3
V4 0 0 15 81 4
V5 0 0 9 81 5
V6 0 0 72 81 6
V7 0 0 0 81 7
V8 0 0 16 81 8
V9 0 0 0 81 9
V10 0 0 16 81 10Your turn
Calculate for each year, the mean abundance of Pocillopora damicornis
# A tibble: 6 x 2
time MeanAbundance
<fct> <dbl>
1 81 30.0
2 83 0.
3 84 0.
4 85 0.
5 87 1.80
6 88 4.00Your turn
Calculate for each year, the number of samples as well as the mean and variance of ozone
lat long month year cloudhigh cloudlow cloudmid ozone pressure surftemp temperature
1 36.20000 -113.8 1 1995 26.0 7.5 34.5 304 835 272.7 272.1
2 33.70435 -113.8 1 1995 20.0 11.5 32.5 304 940 279.5 282.2
3 31.20870 -113.8 1 1995 16.0 16.5 26.0 298 960 284.7 285.2
4 28.71304 -113.8 1 1995 13.0 20.5 14.5 276 990 289.3 290.7
5 26.21739 -113.8 1 1995 7.5 26.0 10.5 274 1000 292.2 292.7
6 23.72174 -113.8 1 1995 8.0 30.0 9.5 264 1000 294.1 293.6Your turn
Calculate for each year, the number of samples as well as the mean and variance of ozone
# A tibble: 6 x 4
year N Mean Var
<int> <int> <dbl> <dbl>
1 1995 6912 264. 258.
2 1996 6912 267. 326.
3 1997 6912 266. 327.
4 1998 6912 267. 507.
5 1999 6912 270. 368.
6 2000 6912 269. 353.Reshaping data
Reshaping data frames
Wide data
| Between | Plot | Time.0 | Time.1 | Time.2 | |
|---|---|---|---|---|---|
| R1 | A1 | P1 | 8 | 14 | 14 |
| R2 | A1 | P2 | 10 | 12 | 11 |
| R3 | A2 | P3 | 7 | 11 | 8 |
| R4 | A2 | P4 | 11 | 9 | 2 |
Wide to long (melt)
> data.w %>% gather(Time,Count,Time.0:Time.2)
> ## OR
> data.w %>% gather(Time,Count, -Between, -Plot) Between Plot Time Count
1 A1 P1 Time.0 8
2 A1 P2 Time.0 10
3 A2 P3 Time.0 7
4 A2 P4 Time.0 11
5 A1 P1 Time.1 14
6 A1 P2 Time.1 12
7 A2 P3 Time.1 11
8 A2 P4 Time.1 9
9 A1 P1 Time.2 14
10 A1 P2 Time.2 11
11 A2 P3 Time.2 8
12 A2 P4 Time.2 2
Between Plot Time Count
1 A1 P1 Time.0 8
2 A1 P2 Time.0 10
3 A2 P3 Time.0 7
4 A2 P4 Time.0 11
5 A1 P1 Time.1 14
6 A1 P2 Time.1 12
7 A2 P3 Time.1 11
8 A2 P4 Time.1 9
9 A1 P1 Time.2 14
10 A1 P2 Time.2 11
11 A2 P3 Time.2 8
12 A2 P4 Time.2 2Reshaping data frames
Long data
| Resp1 | Resp2 | Between | Plot | Subplot | Within |
|---|---|---|---|---|---|
| 8 | 17 | A1 | P1 | S1 | B1 |
| 10 | 18 | A1 | P1 | S1 | B2 |
| 7 | 17 | A1 | P1 | S2 | B1 |
| 11 | 21 | A1 | P1 | S2 | B2 |
| 14 | 19 | A2 | P2 | S3 | B1 |
| 12 | 13 | A2 | P2 | S3 | B2 |
| 11 | 24 | A2 | P2 | S4 | B1 |
| 9 | 18 | A2 | P2 | S4 | B2 |
| 14 | 25 | A3 | P3 | S5 | B1 |
| 11 | 18 | A3 | P3 | S5 | B2 |
| 8 | 27 | A3 | P3 | S6 | B1 |
| 2 | 22 | A3 | P3 | S6 | B2 |
| 8 | 17 | A1 | P4 | S7 | B1 |
| 10 | 22 | A1 | P4 | S7 | B2 |
| 7 | 16 | A1 | P4 | S8 | B1 |
| 12 | 13 | A1 | P4 | S8 | B2 |
| 11 | 23 | A2 | P5 | S9 | B1 |
| 12 | 19 | A2 | P5 | S9 | B2 |
| 12 | 23 | A2 | P5 | S10 | B1 |
| 10 | 21 | A2 | P5 | S10 | B2 |
| 3 | 17 | A3 | P6 | S11 | B1 |
| 11 | 16 | A3 | P6 | S11 | B2 |
| 13 | 26 | A3 | P6 | S12 | B1 |
| 7 | 28 | A3 | P6 | S12 | B2 |
Reshaping data frames
Resp1 Resp2 Between Plot Subplot Within
1 8 17 A1 P1 S1 B1
2 10 18 A1 P1 S1 B2Widen (cast)
Widen Resp1 for repeated measures (Within)
Between Plot Subplot B1 B2
1 A1 P1 S1 8 10
2 A1 P1 S2 7 11
3 A1 P4 S7 8 10
4 A1 P4 S8 7 12
5 A2 P2 S3 14 12
6 A2 P2 S4 11 9
7 A2 P5 S9 11 12
8 A2 P5 S10 12 10
9 A3 P3 S5 14 11
10 A3 P3 S6 8 2
11 A3 P6 S11 3 11
12 A3 P6 S12 13 7Reshaping data frames
Widen Resp1 and Resp2 for repeated measures (Within)
Resp1 Resp2 Between Plot Subplot Within
1 8 17 A1 P1 S1 B1
2 10 18 A1 P1 S1 B2 Between Plot Subplot Within Resp Count
1 A1 P1 S1 B1 Resp1 8
2 A1 P1 S1 B2 Resp1 10
3 A1 P1 S2 B1 Resp1 7
4 A1 P1 S2 B2 Resp1 11
5 A2 P2 S3 B1 Resp1 14
6 A2 P2 S3 B2 Resp1 12
7 A2 P2 S4 B1 Resp1 11
8 A2 P2 S4 B2 Resp1 9
9 A3 P3 S5 B1 Resp1 14
10 A3 P3 S5 B2 Resp1 11
11 A3 P3 S6 B1 Resp1 8
12 A3 P3 S6 B2 Resp1 2
13 A1 P4 S7 B1 Resp1 8
14 A1 P4 S7 B2 Resp1 10
15 A1 P4 S8 B1 Resp1 7
16 A1 P4 S8 B2 Resp1 12
17 A2 P5 S9 B1 Resp1 11
18 A2 P5 S9 B2 Resp1 12
19 A2 P5 S10 B1 Resp1 12
20 A2 P5 S10 B2 Resp1 10
21 A3 P6 S11 B1 Resp1 3
22 A3 P6 S11 B2 Resp1 11
23 A3 P6 S12 B1 Resp1 13
24 A3 P6 S12 B2 Resp1 7
25 A1 P1 S1 B1 Resp2 17
26 A1 P1 S1 B2 Resp2 18
27 A1 P1 S2 B1 Resp2 17
28 A1 P1 S2 B2 Resp2 21
29 A2 P2 S3 B1 Resp2 19
30 A2 P2 S3 B2 Resp2 13
31 A2 P2 S4 B1 Resp2 24
32 A2 P2 S4 B2 Resp2 18
33 A3 P3 S5 B1 Resp2 25
34 A3 P3 S5 B2 Resp2 18
35 A3 P3 S6 B1 Resp2 27
36 A3 P3 S6 B2 Resp2 22
37 A1 P4 S7 B1 Resp2 17
38 A1 P4 S7 B2 Resp2 22
39 A1 P4 S8 B1 Resp2 16
40 A1 P4 S8 B2 Resp2 13
41 A2 P5 S9 B1 Resp2 23
42 A2 P5 S9 B2 Resp2 19
43 A2 P5 S10 B1 Resp2 23
44 A2 P5 S10 B2 Resp2 21
45 A3 P6 S11 B1 Resp2 17
46 A3 P6 S11 B2 Resp2 16
47 A3 P6 S12 B1 Resp2 26
48 A3 P6 S12 B2 Resp2 28Reshaping data frames
Widen Resp1 and Resp2 for repeated measures (Within)
Resp1 Resp2 Between Plot Subplot Within
1 8 17 A1 P1 S1 B1
2 10 18 A1 P1 S1 B2 Between Plot Subplot WR Count
1 A1 P1 S1 B1_Resp1 8
2 A1 P1 S1 B2_Resp1 10
3 A1 P1 S2 B1_Resp1 7
4 A1 P1 S2 B2_Resp1 11
5 A2 P2 S3 B1_Resp1 14
6 A2 P2 S3 B2_Resp1 12
7 A2 P2 S4 B1_Resp1 11
8 A2 P2 S4 B2_Resp1 9
9 A3 P3 S5 B1_Resp1 14
10 A3 P3 S5 B2_Resp1 11
11 A3 P3 S6 B1_Resp1 8
12 A3 P3 S6 B2_Resp1 2
13 A1 P4 S7 B1_Resp1 8
14 A1 P4 S7 B2_Resp1 10
15 A1 P4 S8 B1_Resp1 7
16 A1 P4 S8 B2_Resp1 12
17 A2 P5 S9 B1_Resp1 11
18 A2 P5 S9 B2_Resp1 12
19 A2 P5 S10 B1_Resp1 12
20 A2 P5 S10 B2_Resp1 10
21 A3 P6 S11 B1_Resp1 3
22 A3 P6 S11 B2_Resp1 11
23 A3 P6 S12 B1_Resp1 13
24 A3 P6 S12 B2_Resp1 7
25 A1 P1 S1 B1_Resp2 17
26 A1 P1 S1 B2_Resp2 18
27 A1 P1 S2 B1_Resp2 17
28 A1 P1 S2 B2_Resp2 21
29 A2 P2 S3 B1_Resp2 19
30 A2 P2 S3 B2_Resp2 13
31 A2 P2 S4 B1_Resp2 24
32 A2 P2 S4 B2_Resp2 18
33 A3 P3 S5 B1_Resp2 25
34 A3 P3 S5 B2_Resp2 18
35 A3 P3 S6 B1_Resp2 27
36 A3 P3 S6 B2_Resp2 22
37 A1 P4 S7 B1_Resp2 17
38 A1 P4 S7 B2_Resp2 22
39 A1 P4 S8 B1_Resp2 16
40 A1 P4 S8 B2_Resp2 13
41 A2 P5 S9 B1_Resp2 23
42 A2 P5 S9 B2_Resp2 19
43 A2 P5 S10 B1_Resp2 23
44 A2 P5 S10 B2_Resp2 21
45 A3 P6 S11 B1_Resp2 17
46 A3 P6 S11 B2_Resp2 16
47 A3 P6 S12 B1_Resp2 26
48 A3 P6 S12 B2_Resp2 28Reshaping data frames
Widen Resp1 and Resp2 for repeated measures (Within)
Resp1 Resp2 Between Plot Subplot Within
1 8 17 A1 P1 S1 B1
2 10 18 A1 P1 S1 B2 Between Plot Subplot B1_Resp1 B1_Resp2 B2_Resp1 B2_Resp2
1 A1 P1 S1 8 17 10 18
2 A1 P1 S2 7 17 11 21
3 A1 P4 S7 8 17 10 22
4 A1 P4 S8 7 16 12 13
5 A2 P2 S3 14 19 12 13
6 A2 P2 S4 11 24 9 18
7 A2 P5 S9 11 23 12 19
8 A2 P5 S10 12 23 10 21
9 A3 P3 S5 14 25 11 18
10 A3 P3 S6 8 27 2 22
11 A3 P6 S11 3 17 11 16
12 A3 P6 S12 13 26 7 28Combining data
Merging data frames
Bio data (missing Subplot 3)
| Resp1 | Resp2 | Between | Plot | Subplot | |
|---|---|---|---|---|---|
| 1 | 8 | 18 | A1 | P1 | S1 |
| 2 | 10 | 21 | A1 | P1 | S2 |
| 4 | 11 | 23 | A1 | P2 | S4 |
| 5 | 14 | 22 | A2 | P3 | S5 |
| 6 | 12 | 24 | A2 | P3 | S6 |
| 7 | 11 | 23 | A2 | P4 | S7 |
| 8 | 9 | 20 | A2 | P4 | S8 |
| 9 | 14 | 11 | A3 | P5 | S9 |
| 10 | 11 | 22 | A3 | P5 | S10 |
| 11 | 8 | 24 | A3 | P6 | S11 |
| 12 | 2 | 16 | A3 | P6 | S12 |
Physio-chemical data (missing S7)
| Chem1 | Chem2 | Between | Plot | Subplot | |
|---|---|---|---|---|---|
| 1 | 1.453 | 0.8858 | A1 | P1 | S1 |
| 2 | 3.266 | 0.18 | A1 | P1 | S2 |
| 3 | 1.179 | 5.078 | A1 | P2 | S3 |
| 4 | 13.4 | 1.576 | A1 | P2 | S4 |
| 5 | 3.779 | 1.622 | A2 | P3 | S5 |
| 6 | 1.197 | 4.237 | A2 | P3 | S6 |
| 8 | 5.688 | 2.986 | A2 | P4 | S8 |
| 9 | 4.835 | 4.133 | A3 | P5 | S9 |
| 10 | 2.003 | 3.604 | A3 | P5 | S10 |
| 11 | 12.33 | 1.776 | A3 | P6 | S11 |
| 12 | 4.014 | 0.2255 | A3 | P6 | S12 |
Merging data frames
Merge bio and chem data (only keep full matches - an inner join)
Resp1 Resp2 Between Plot Subplot Chem1 Chem2
1 8 18 A1 P1 S1 1.452878 0.8858208
2 10 21 A1 P1 S2 3.266253 0.1800177
3 11 23 A1 P2 S4 13.400350 1.5762780
4 14 22 A2 P3 S5 3.779183 1.6222430
5 12 24 A2 P3 S6 1.196657 4.2369184
6 9 20 A2 P4 S8 5.687807 2.9859003
7 14 11 A3 P5 S9 4.834518 4.1328919
8 11 22 A3 P5 S10 2.002931 3.6043314
9 8 24 A3 P6 S11 12.326867 1.7763576
10 2 16 A3 P6 S12 4.014221 0.2255188- S3 and S7 absent
Merging data frames
Merge bio and chem data (keep all data - outer join)
Resp1 Resp2 Between Plot Subplot Chem1 Chem2
1 8 18 A1 P1 S1 1.452878 0.8858208
2 10 21 A1 P1 S2 3.266253 0.1800177
3 11 23 A1 P2 S4 13.400350 1.5762780
4 14 22 A2 P3 S5 3.779183 1.6222430
5 12 24 A2 P3 S6 1.196657 4.2369184
6 11 23 A2 P4 S7 NA NA
7 9 20 A2 P4 S8 5.687807 2.9859003
8 14 11 A3 P5 S9 4.834518 4.1328919
9 11 22 A3 P5 S10 2.002931 3.6043314
10 8 24 A3 P6 S11 12.326867 1.7763576
11 2 16 A3 P6 S12 4.014221 0.2255188
12 NA NA A1 P2 S3 1.178652 5.0780682- note the order of Subplot
Merging data frames
Merge bio and chem data (only keep full BIO matches - left join)
Resp1 Resp2 Between Plot Subplot Chem1 Chem2
1 8 18 A1 P1 S1 1.452878 0.8858208
2 10 21 A1 P1 S2 3.266253 0.1800177
3 11 23 A1 P2 S4 13.400350 1.5762780
4 14 22 A2 P3 S5 3.779183 1.6222430
5 12 24 A2 P3 S6 1.196657 4.2369184
6 11 23 A2 P4 S7 NA NA
7 9 20 A2 P4 S8 5.687807 2.9859003
8 14 11 A3 P5 S9 4.834518 4.1328919
9 11 22 A3 P5 S10 2.002931 3.6043314
10 8 24 A3 P6 S11 12.326867 1.7763576
11 2 16 A3 P6 S12 4.014221 0.2255188Merging data frames
Merge bio and chem data (only keep full CHEM matches - right join)
Resp1 Resp2 Between Plot Subplot Chem1 Chem2
1 8 18 A1 P1 S1 1.452878 0.8858208
2 10 21 A1 P1 S2 3.266253 0.1800177
3 NA NA A1 P2 S3 1.178652 5.0780682
4 11 23 A1 P2 S4 13.400350 1.5762780
5 14 22 A2 P3 S5 3.779183 1.6222430
6 12 24 A2 P3 S6 1.196657 4.2369184
7 9 20 A2 P4 S8 5.687807 2.9859003
8 14 11 A3 P5 S9 4.834518 4.1328919
9 11 22 A3 P5 S10 2.002931 3.6043314
10 8 24 A3 P6 S11 12.326867 1.7763576
11 2 16 A3 P6 S12 4.014221 0.2255188VLOOKUP
VLOOKUP
Biological data set (data.bio)
Resp1 Resp2 Between Plot Subplot
1 8 18 A1 P1 S1
2 10 21 A1 P1 S2
4 11 23 A1 P2 S4
5 14 22 A2 P3 S5
6 12 24 A2 P3 S6
7 11 23 A2 P4 S7
8 9 20 A2 P4 S8
9 14 11 A3 P5 S9
10 11 22 A3 P5 S10
11 8 24 A3 P6 S11
12 2 16 A3 P6 S12Geographical data set (lookup table) (data.geo)
Plot LAT LONG
1 P1 17.9605 145.4326
2 P2 17.5210 146.1983
3 P3 17.0011 146.3839
4 P4 18.2350 146.7934
5 P5 18.9840 146.0345
6 P6 20.1154 146.4672VLOOKUP
Incorporate (merge) the lat/longs into the bio data
Resp1 Resp2 Between Plot Subplot LAT LONG
1 8 18 A1 P1 S1 17.9605 145.4326
2 10 21 A1 P1 S2 17.9605 145.4326
3 11 23 A1 P2 S4 17.5210 146.1983
4 14 22 A2 P3 S5 17.0011 146.3839
5 12 24 A2 P3 S6 17.0011 146.3839
6 11 23 A2 P4 S7 18.2350 146.7934
7 9 20 A2 P4 S8 18.2350 146.7934
8 14 11 A3 P5 S9 18.9840 146.0345
9 11 22 A3 P5 S10 18.9840 146.0345
10 8 24 A3 P6 S11 20.1154 146.4672
11 2 16 A3 P6 S12 20.1154 146.4672Applied examples
Tikus Island coral data
Psammocora contigua Psammocora digitata time rep
1 0 0 81 1
2 0 0 81 2
3 0 0 81 3
4 0 0 81 4
5 0 0 81 5
6 0 0 81 6
7 0 0 81 7
8 0 0 81 8
9 0 0 81 9
10 0 0 81 10Observations: 60
Variables: 77
$ `Psammocora contigua` <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
$ `Psammocora digitata` <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
$ `Pocillopora damicornis` <int> 79, 51, 42, 15, 9, 72, 0, 16, 0,...
$ `Pocillopora verrucosa` <int> 32, 21, 35, 0, 0, 0, 41, 25, 38,...
$ `Stylopora pistillata` <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
$ `Acropora bruegemanni` <int> 0, 44, 0, 11, 9, 10, 0, 0, 0, 37...
$ `Acropora robusta` <int> 0, 35, 40, 0, 0, 0, 0, 0, 0, 0, ...
$ `Acropora grandis` <int> 0, 0, 0, 0, 0, 0, 60, 0, 0, 0, 0...
$ `Acropora intermedia` <int> 30, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0...
$ `Acropora formosa` <int> 75, 0, 15, 0, 125, 0, 0, 0, 10, ...
$ `Acropora splendida` <int> 0, 22, 0, 31, 0, 9, 16, 0, 0, 20...
$ `Acropera aspera` <int> 17, 18, 9, 8, 23, 0, 17, 13, 16,...
$ `Acropora hyacinthus` <int> 141, 34, 55, 54, 0, 0, 0, 0, 0, ...
$ `Acropora palifera` <int> 32, 0, 44, 0, 17, 0, 0, 0, 0, 0,...
$ `Acropora cytherea` <int> 108, 33, 14, 122, 0, 0, 0, 8, 0,...
$ `Acropora tenuis` <int> 0, 25, 0, 0, 0, 22, 28, 0, 0, 0,...
$ `Acropora pulchra` <int> 0, 0, 15, 52, 62, 33, 0, 0, 24, ...
$ `Acropora nasuta` <int> 43, 21, 19, 0, 0, 0, 10, 0, 0, 0...
$ `Acropora humilis` <int> 31, 25, 0, 19, 0, 0, 0, 0, 0, 0,...
$ `Acropora diversa` <int> 22, 19, 20, 13, 23, 14, 0, 12, 1...
$ `Acropora digitifera` <int> 30, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0...
$ `Acropora divaricata` <int> 0, 32, 55, 0, 0, 0, 0, 0, 0, 0, ...
$ `Acropora subglabra` <int> 51, 0, 0, 44, 15, 0, 0, 25, 0, 0...
$ `Acropora cerealis` <int> 0, 75, 0, 0, 0, 0, 0, 0, 0, 0, 0...
$ `Acropora valida` <int> 0, 0, 0, 30, 0, 0, 0, 0, 0, 0, 0...
$ `Acropora acuminata` <int> 20, 0, 71, 0, 15, 0, 25, 25, 0, ...
$ `Acropora elsevi` <int> 30, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0...
$ `Acropora millepora` <int> 17, 14, 0, 20, 0, 0, 0, 0, 0, 0,...
$ `Montipora monasteriata` <int> 60, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0...
$ `Montipora tuberculosa` <int> 0, 15, 15, 0, 0, 0, 0, 0, 0, 0, ...
$ `Montipora hispida` <int> 0, 0, 0, 32, 40, 24, 0, 0, 0, 0,...
$ `Montipora digitata` <int> 0, 0, 0, 0, 0, 77, 84, 53, 71, 3...
$ `Montipora foliosa` <int> 0, 0, 0, 0, 50, 71, 62, 81, 24, ...
$ `Montipora verrucosa` <int> 0, 0, 30, 0, 0, 0, 0, 0, 0, 0, 0...
$ `Fungia fungites` <int> 0, 0, 18, 17, 0, 0, 0, 0, 0, 0, ...
$ `Fungia paumotensis` <int> 0, 33, 0, 0, 0, 0, 0, 0, 0, 0, 1...
$ `Fungia concina` <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
$ `Fungia scutaria` <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
$ `Halomitra limax` <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
$ `Pavona varians` <int> 30, 0, 0, 0, 0, 0, 0, 0, 0, 0, 3...
$ `Pavona venosa` <int> 0, 24, 0, 0, 0, 0, 0, 0, 0, 0, 0...
$ `Pavona cactus` <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
$ `Coeloseris mayeri` <int> 20, 0, 15, 0, 9, 19, 0, 0, 25, 0...
$ `Galaxea fascicularis` <int> 51, 27, 31, 24, 0, 13, 0, 0, 0, ...
$ `Symphyllia radians` <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
$ `Lobophyllia corymbosa` <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
$ `Lobophyllia hemprichii` <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
$ `Porites cylindrica` <int> 61, 24, 0, 20, 0, 0, 0, 0, 0, 0,...
$ `Porites lichen` <int> 0, 47, 49, 0, 0, 0, 0, 0, 0, 0, ...
$ `Porites lobata` <int> 36, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0...
$ `Porites lutea` <int> 30, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0...
$ `Porites nigrescens` <int> 0, 0, 0, 21, 0, 9, 25, 0, 45, 26...
$ `Porites solida` <int> 0, 0, 10, 0, 17, 0, 31, 41, 0, 0...
$ `Porites stephensoni` <int> 0, 0, 0, 0, 0, 0, 0, 30, 0, 0, 0...
$ `Goniopora lobata` <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
$ `Favia pallida` <int> 10, 20, 0, 0, 0, 0, 0, 0, 0, 0, ...
$ `Favia speciosa` <int> 0, 0, 30, 0, 0, 0, 0, 0, 0, 0, 0...
$ `Favia stelligera` <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
$ `Favia rotumana` <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
$ `Favites abdita` <int> 33, 41, 23, 27, 91, 63, 72, 48, ...
$ `Favites chinensis` <int> 0, 44, 78, 61, 44, 0, 55, 30, 30...
$ `Goniastrea rectiformis` <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 6,...
$ `Goniastrea pectinata` <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
$ `Goniastrea sp` <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
$ `Dulophyllia crispa` <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
$ `Platygyra daedalea` <int> 0, 27, 55, 0, 71, 74, 55, 48, 0,...
$ `Platygyra sinensis` <int> 47, 27, 56, 26, 0, 0, 0, 0, 0, 0...
$ `Hydnopora rigida` <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
$ `Leptastrea purpurea` <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
$ `Leptastrea pruinosa` <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
$ `Cyphastrea serailia` <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 19...
$ `Millepora platyphylla` <int> 30, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0...
$ `Millepora dichotoma` <int> 21, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0...
$ `Millepora intrincata` <int> 24, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0...
$ `Heliopora coerulea` <int> 461, 271, 221, 154, 0, 0, 0, 0, ...
$ time <fct> 81, 81, 81, 81, 81, 81, 81, 81, ...
$ rep <fct> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 1...Tikus Island coral data
Explore/Process data
- Convert abundance to cover
- Mean cover of total Acropora per year
- NOTE there is a typo ‘Acropera’
Tikus Island coral data
Explore/Process data
- Convert abundance to cover (abundance is the length in cm of a 10m transect containing the species)
- Mean cover of total Acropora per year
- NOTE there is a typo ‘Acropera’
Step 1. fix typo (rename) - backticks
Tikus Island coral data
Explore/Process data
- Convert abundance to cover
- Mean cover of total Acropora per year
- NOTE there is a typo ‘Acropera’
Step 2. melt data (gather)
Tikus Island coral data
Explore/Process data
- Convert abundance to cover
- Mean cover of total Acropora per year
- NOTE there is a typo ‘Acropera’
Step 3. Calculate Cover (mutate) (Abundance/10)
Tikus Island coral data
Explore/Process data
- Convert abundance to cover
- Mean cover of total Acropora per year
- NOTE there is a typo ‘Acropera’
Step 4. Split species into Genera and Species (separate)
Tikus Island coral data
Explore/Process data
- Convert abundance to cover
- Mean cover of total Acropora per year
- NOTE there is a typo ‘Acropera’
Step 5. Subset just ‘Acropora’ (filter)
Tikus Island coral data
Explore/Process data
- Convert abundance to cover
- Mean cover of total Acropora per year
- NOTE there is a typo ‘Acropera’
Step 6. Sum over all Species (group_by and summarise)
Tikus Island coral data
Explore/Process data
- Convert abundance to cover
- Mean cover of total Acropora per year
- NOTE there is a typo ‘Acropera’
Step 7. Summarise per year
> tikus %>% rename(`Acropora aspera`=`Acropera aspera`) %>%
+ gather(Species, Abundance,-time,-rep) %>%
+ mutate(Cover=Abundance/10) %>%
+ separate(Species,c('Genera','Species')) %>%
+ filter(Genera=='Acropora') %>%
+ group_by(time,rep) %>%
+ summarise(SumCover=sum(Cover)) %>%
+ group_by(time) %>%
+ summarise(Mean=mean(SumCover),
+ Var=var(SumCover))# A tibble: 6 x 3
time Mean Var
<fct> <dbl> <dbl>
1 81 25.6 383.
2 83 0. 0.
3 84 0. 0.
4 85 2.43 14.2
5 87 8.01 68.5
6 88 8.55 106. Tikus Island coral data
> tikus %>% rename(`Acropora aspera`=`Acropera aspera`) %>%
+ gather(Species, Abundance,-time,-rep) %>%
+ mutate(Cover=Abundance/10) %>%
+ separate(Species,c('Genera','Species')) %>%
+ filter(Genera=='Acropora') %>%
+ group_by(time,rep) %>%
+ summarise(SumCover=sum(Cover)) %>%
+ group_by(time) %>%
+ summarise(Mean=mean(SumCover),
+ Var=var(SumCover))# A tibble: 6 x 3
time Mean Var
<fct> <dbl> <dbl>
1 81 25.6 383.
2 83 0. 0.
3 84 0. 0.
4 85 2.43 14.2
5 87 8.01 68.5
6 88 8.55 106. Tikus Island coral data
Can you modify so that we get the means and var for each Genera per year?
> tikus %>% rename(`Acropora aspera`=`Acropera aspera`) %>%
+ gather(Species, Abundance,-time,-rep) %>%
+ mutate(Cover=Abundance/10) %>%
+ separate(Species,c('Genera','Species')) %>%
+ group_by(time,rep,Genera) %>%
+ summarise(SumCover=sum(Cover)) %>%
+ group_by(time,Genera) %>%
+ summarise(Mean=mean(SumCover),
+ Var=var(SumCover))# A tibble: 144 x 4
# Groups: time [?]
time Genera Mean Var
<fct> <chr> <dbl> <dbl>
1 81 Acropora 25.6 383.
2 81 Coeloseris 0.880 1.02
3 81 Cyphastrea 0. 0.
4 81 Dulophyllia 0. 0.
5 81 Favia 0.600 1.16
6 81 Favites 8.22 14.9
7 81 Fungia 0.680 1.38
8 81 Galaxea 1.46 3.23
9 81 Goniastrea 0. 0.
10 81 Goniopora 0. 0.
# ... with 134 more rowsTikus Island coral data
What about the means and var for the top 3 Genera per year (sorted from highest to lowest)?
> tikus %>% rename(`Acropora aspera`=`Acropera aspera`) %>%
+ gather(Species, Abundance,-time,-rep) %>%
+ mutate(Cover=Abundance/10) %>%
+ separate(Species,c('Genera','Species')) %>%
+ group_by(time,rep,Genera) %>%
+ summarise(SumCover=sum(Cover)) %>%
+ group_by(time,Genera) %>%
+ summarise(Mean=mean(SumCover),
+ Var=var(SumCover)) %>%
+ top_n(3,Mean) %>%
+ arrange(desc(Mean))# A tibble: 18 x 4
# Groups: time [6]
time Genera Mean Var
<fct> <chr> <dbl> <dbl>
1 87 Montipora 27.4 966.
2 81 Acropora 25.6 383.
3 85 Montipora 20.5 171.
4 85 Porites 19.0 51.3
5 88 Montipora 11.8 644.
6 81 Montipora 11.4 95.7
7 81 Heliopora 11.1 262.
8 84 Montipora 11.0 70.5
9 88 Porites 9.84 41.4
10 88 Acropora 8.55 106.
11 87 Acropora 8.01 68.5
12 87 Porites 4.49 35.8
13 84 Porites 2.94 6.65
14 85 Platygyra 2.55 8.74
15 83 Porites 1.74 2.07
16 84 Pavona 1.20 3.33
17 83 Fungia 1.14 3.64
18 83 Montipora 0.930 1.57