ggplot
> library(ggplot2)
> library(grid)
> library(gridExtra)
> library(scales)> head(BOD) Time demand
1 1 8.3
2 2 10.3
3 3 19.0
4 4 16.0
5 5 15.6
6 7 19.8> summary(BOD) Time demand
Min. :1.000 Min. : 8.30
1st Qu.:2.250 1st Qu.:11.62
Median :3.500 Median :15.80
Mean :3.667 Mean :14.83
3rd Qu.:4.750 3rd Qu.:18.25
Max. :7.000 Max. :19.80 > p <- ggplot() +
+ #single layer - points
+ layer(data=BOD, #data.frame
+ mapping=aes(y=demand,x=Time),
+ stat="identity", #use original data
+ geom="point", #plot data as points
+ position="identity",
+ params = list(na.rm = TRUE),
+ show.legend = FALSE
+ )+ #layer of lines
+ layer( data=BOD, #data.frame
+ mapping=aes(y=demand,x=Time),
+ stat="identity", #use original data
+ geom="line", #plot data as a line
+ position="identity",
+ params = list(na.rm = TRUE),
+ show.legend = FALSE
+ ) +
+ coord_cartesian() + #cartesian coordinates
+ scale_x_continuous() + #continuous x axis
+ scale_y_continuous() #continuous y axis
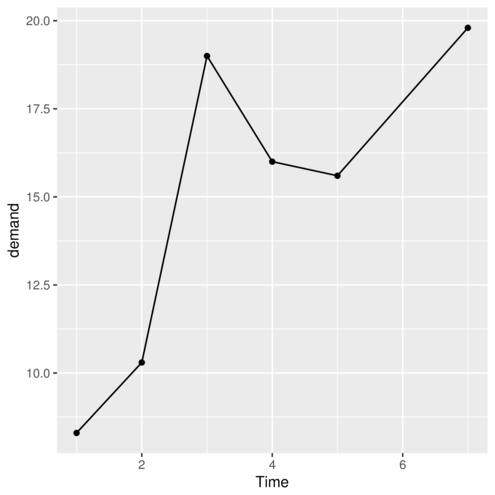
> p #print the plot
> ggplot(data=BOD, map=aes(y=demand,x=Time)) + geom_point()+geom_line()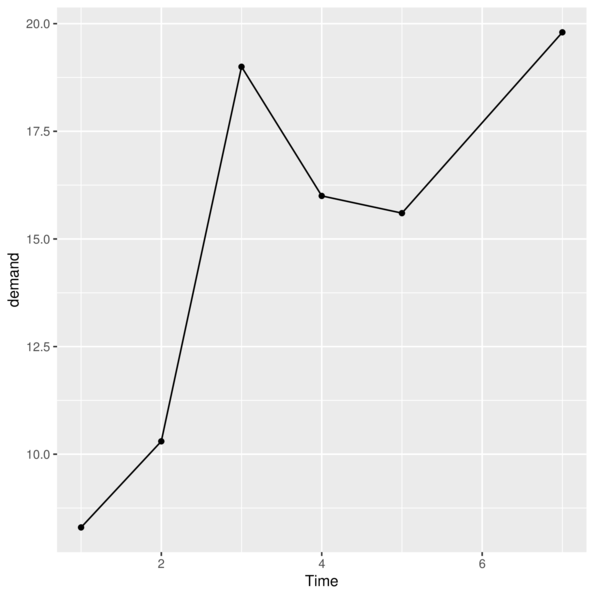
> p<-ggplot(data=BOD)> p<-p + geom_point(aes(y=demand, x=Time))
> p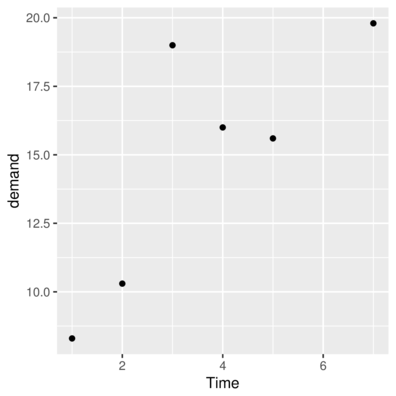
> p<-ggplot(data=BOD)> p<-p + geom_point(aes(y=demand, x=Time))> p <- p + scale_x_sqrt(name="Time")
> p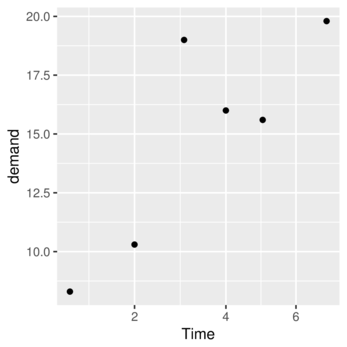
geom_ and stat_stat_identitygeom_If omitted, inherited from ggplot()
stat_ functiongeom_> ggplot(data=BOD, aes(y=demand, x=Time)) + geom_point()
> #OR
> ggplot(data=BOD) + geom_point(aes(y=demand, x=Time))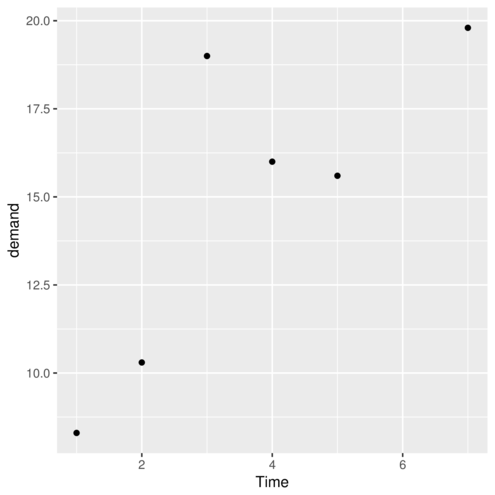
geom_point> head(CO2) Plant Type Treatment conc uptake
1 Qn1 Quebec nonchilled 95 16.0
2 Qn1 Quebec nonchilled 175 30.4
3 Qn1 Quebec nonchilled 250 34.8
4 Qn1 Quebec nonchilled 350 37.2
5 Qn1 Quebec nonchilled 500 35.3
6 Qn1 Quebec nonchilled 675 39.2> summary(CO2) Plant Type Treatment conc uptake
Qn1 : 7 Quebec :42 nonchilled:42 Min. : 95 Min. : 7.70
Qn2 : 7 Mississippi:42 chilled :42 1st Qu.: 175 1st Qu.:17.90
Qn3 : 7 Median : 350 Median :28.30
Qc1 : 7 Mean : 435 Mean :27.21
Qc3 : 7 3rd Qu.: 675 3rd Qu.:37.12
Qc2 : 7 Max. :1000 Max. :45.50
(Other):42 geom_point> ggplot(CO2)+geom_point(aes(x=conc,y=uptake), colour="red")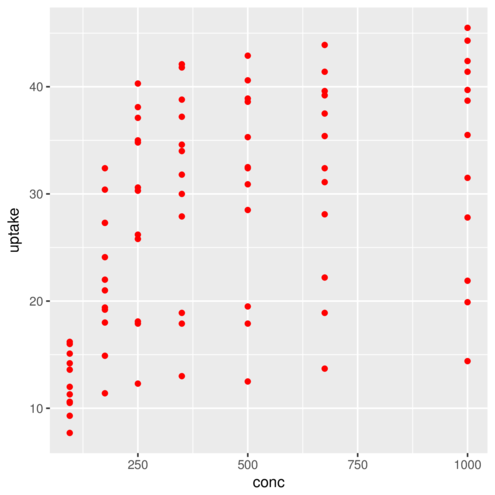
geom_point> ggplot(CO2)+geom_point(aes(x=conc,y=uptake, colour=Type))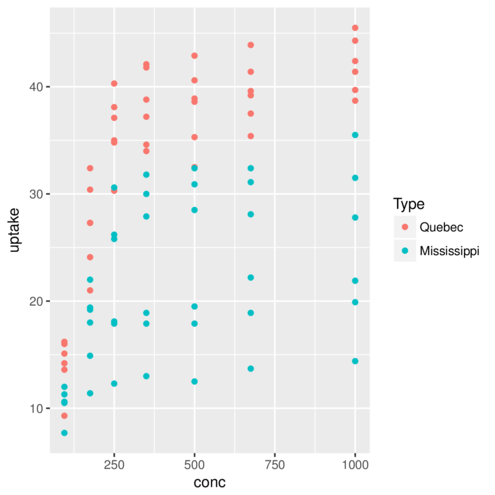
geom_point> ggplot(CO2)+geom_point(aes(x=conc,y=uptake),
+ stat="summary",fun.y=mean)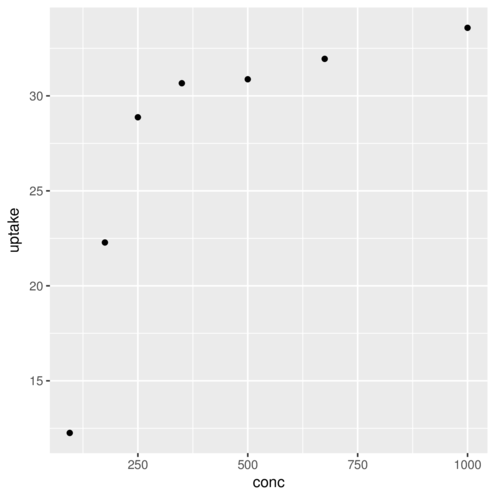
> head(diamonds)# A tibble: 6 x 10
carat cut color clarity depth table price x y z
<dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
1 0.23 Ideal E SI2 61.5 55 326 3.95 3.98 2.43
2 0.21 Premium E SI1 59.8 61 326 3.89 3.84 2.31
3 0.23 Good E VS1 56.9 65 327 4.05 4.07 2.31
4 0.29 Premium I VS2 62.4 58 334 4.20 4.23 2.63
5 0.31 Good J SI2 63.3 58 335 4.34 4.35 2.75
6 0.24 Very Good J VVS2 62.8 57 336 3.94 3.96 2.48> summary(diamonds) carat cut color clarity depth table
Min. :0.2000 Fair : 1610 D: 6775 SI1 :13065 Min. :43.00 Min. :43.00
1st Qu.:0.4000 Good : 4906 E: 9797 VS2 :12258 1st Qu.:61.00 1st Qu.:56.00
Median :0.7000 Very Good:12082 F: 9542 SI2 : 9194 Median :61.80 Median :57.00
Mean :0.7979 Premium :13791 G:11292 VS1 : 8171 Mean :61.75 Mean :57.46
3rd Qu.:1.0400 Ideal :21551 H: 8304 VVS2 : 5066 3rd Qu.:62.50 3rd Qu.:59.00
Max. :5.0100 I: 5422 VVS1 : 3655 Max. :79.00 Max. :95.00
J: 2808 (Other): 2531
price x y z
Min. : 326 Min. : 0.000 Min. : 0.000 Min. : 0.000
1st Qu.: 950 1st Qu.: 4.710 1st Qu.: 4.720 1st Qu.: 2.910
Median : 2401 Median : 5.700 Median : 5.710 Median : 3.530
Mean : 3933 Mean : 5.731 Mean : 5.735 Mean : 3.539
3rd Qu.: 5324 3rd Qu.: 6.540 3rd Qu.: 6.540 3rd Qu.: 4.040
Max. :18823 Max. :10.740 Max. :58.900 Max. :31.800
geom_bar| Feature | geom | stat | position |
|---|---|---|---|
| Histogram | _bar |
_bin |
stack |
> ggplot(diamonds) + geom_bar(aes(x = carat))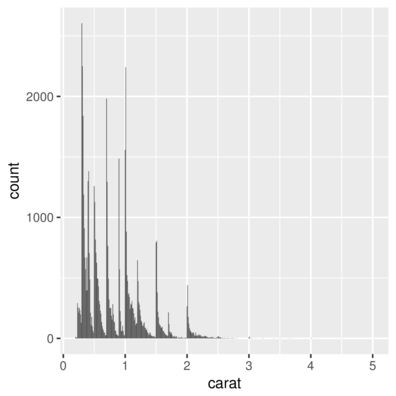
geom_bar| Feature | geom | stat | position |
|---|---|---|---|
| Barchart | _bar |
_bin |
stack |
> ggplot(diamonds) + geom_bar(aes(x = cut))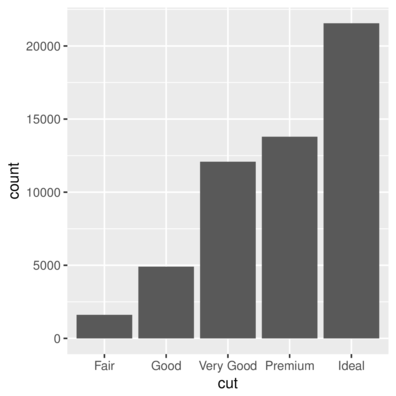
geom_bar| Feature | geom | stat | position |
|---|---|---|---|
| barchart | _bar |
_bin |
stack |
> ggplot(diamonds) + geom_bar(aes(x = cut, fill = clarity))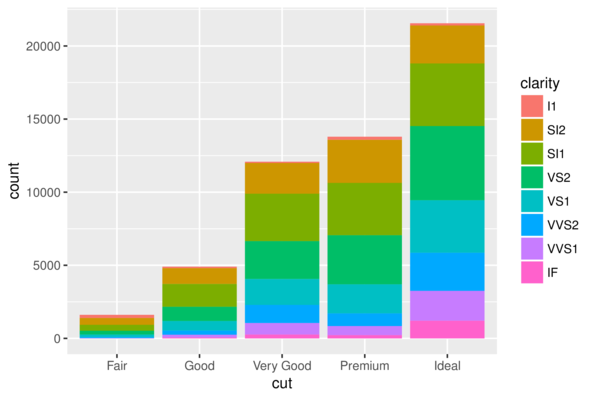
geom_bar| Feature | geom | stat | position |
|---|---|---|---|
| barchart | _bar |
_bin |
stack |
> ggplot(diamonds) + geom_bar(aes(x = cut, fill = clarity))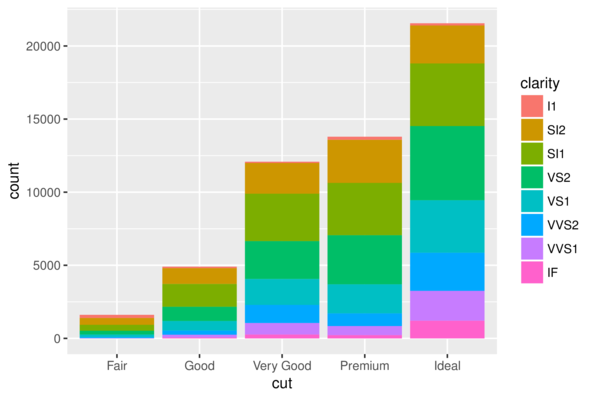
geom_bar| Feature | geom | stat | position |
|---|---|---|---|
| barchart | _bar |
_bin |
dodge |
> ggplot(diamonds) + geom_bar(aes(x = cut, fill = clarity),
+ position='dodge')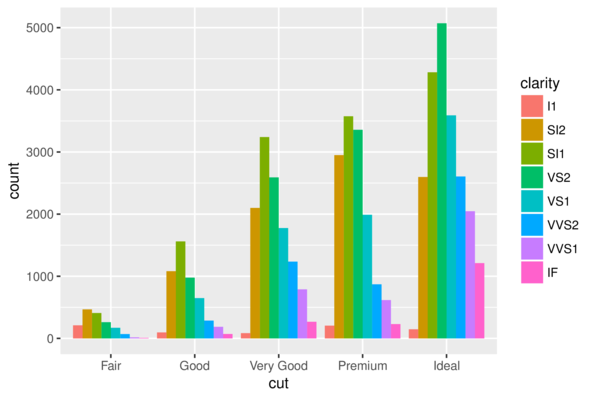
geom_boxplot| Feature | geom | stat | position |
|---|---|---|---|
| boxplot | _boxplot |
_boxplot |
dodge |
> ggplot(diamonds) + geom_boxplot(aes(x = "carat", y = carat))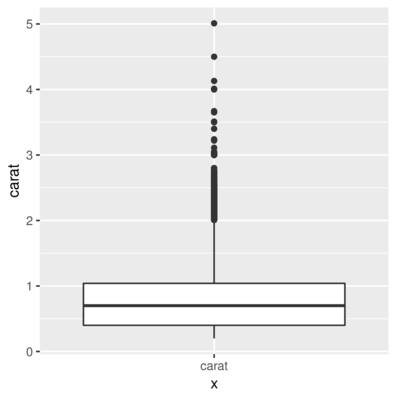
geom_boxplot| Feature | geom | stat | position |
|---|---|---|---|
| boxplot | _boxplot |
_boxplot |
dodge |
> ggplot(diamonds) + geom_boxplot(aes(x = cut, y = carat))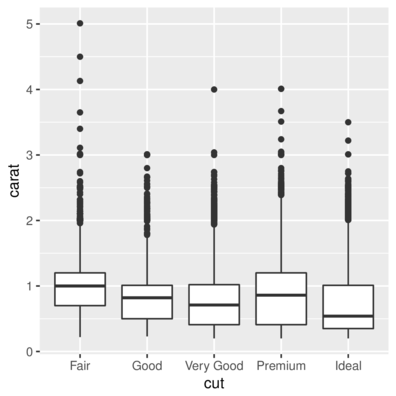
geom_line| Feature | geom | stat | position |
|---|---|---|---|
| line | _line |
_identity |
identity |
> head(CO2, 3) Plant Type Treatment conc uptake
1 Qn1 Quebec nonchilled 95 16.0
2 Qn1 Quebec nonchilled 175 30.4
3 Qn1 Quebec nonchilled 250 34.8> ggplot(CO2) + geom_line(aes(x = conc, y = uptake))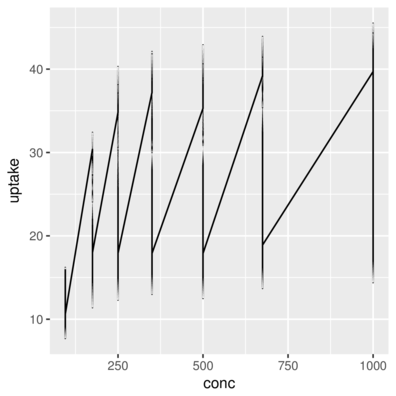
geom_line| Feature | geom | stat | position |
|---|---|---|---|
| line | _line |
_identity |
identity |
> head(CO2, 3) Plant Type Treatment conc uptake
1 Qn1 Quebec nonchilled 95 16.0
2 Qn1 Quebec nonchilled 175 30.4
3 Qn1 Quebec nonchilled 250 34.8> ggplot(CO2) + geom_line(aes(x = conc, y = uptake, group=Plant))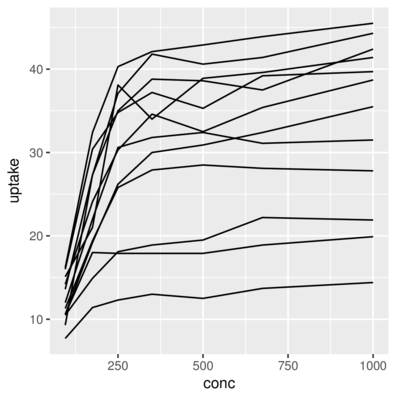
geom_line| Feature | geom | stat | position |
|---|---|---|---|
| line | _line |
_identity |
identity |
> head(CO2, 3) Plant Type Treatment conc uptake
1 Qn1 Quebec nonchilled 95 16.0
2 Qn1 Quebec nonchilled 175 30.4
3 Qn1 Quebec nonchilled 250 34.8> ggplot(CO2) + geom_line(aes(x = conc, y = uptake, color=Plant))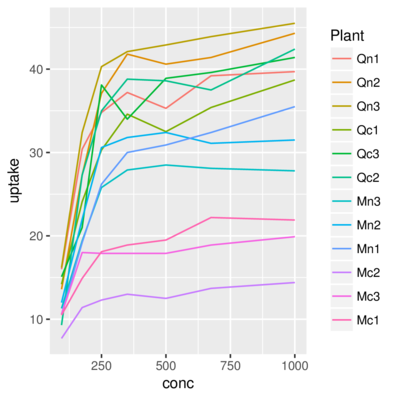
geom_line| Feature | geom | stat | position |
|---|---|---|---|
| line | _line |
_summary |
identity |
> head(CO2, 3) Plant Type Treatment conc uptake
1 Qn1 Quebec nonchilled 95 16.0
2 Qn1 Quebec nonchilled 175 30.4
3 Qn1 Quebec nonchilled 250 34.8> ggplot(CO2) + geom_line(aes(x = conc, y = uptake),
+ stat = "summary", fun.y = mean, color='blue')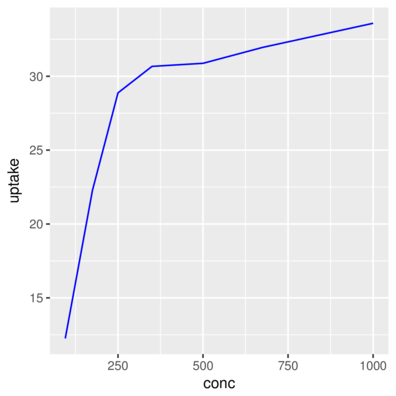
geom_point| Feature | geom | stat | position |
|---|---|---|---|
| point | _point |
_identity |
identity |
> head(CO2, 3) Plant Type Treatment conc uptake
1 Qn1 Quebec nonchilled 95 16.0
2 Qn1 Quebec nonchilled 175 30.4
3 Qn1 Quebec nonchilled 250 34.8> ggplot(CO2) + geom_point(aes(x = conc, y = uptake))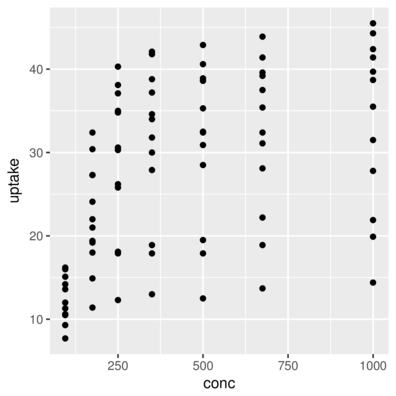
geom_point| Feature | geom | stat | position |
|---|---|---|---|
| point | _point |
_identity |
identity |
> head(CO2, 3) Plant Type Treatment conc uptake
1 Qn1 Quebec nonchilled 95 16.0
2 Qn1 Quebec nonchilled 175 30.4
3 Qn1 Quebec nonchilled 250 34.8> ggplot(CO2) + geom_point(aes(x = conc, y = uptake, fill=Treatment),
+ shape=21)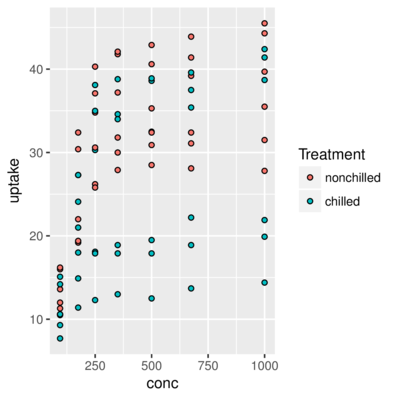
geom_smooth| Feature | geom | stat | position |
|---|---|---|---|
| smoother | _smooth |
_smooth |
identity |
> head(CO2, 3) Plant Type Treatment conc uptake
1 Qn1 Quebec nonchilled 95 16.0
2 Qn1 Quebec nonchilled 175 30.4
3 Qn1 Quebec nonchilled 250 34.8> ggplot(CO2) + geom_smooth(aes(x = conc, y = uptake), method='lm')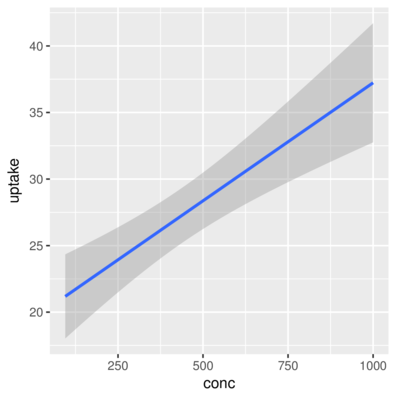
geom_smooth| Feature | geom | stat | position |
|---|---|---|---|
| smoother | _smooth |
_smooth |
identity |
> head(CO2, 3) Plant Type Treatment conc uptake
1 Qn1 Quebec nonchilled 95 16.0
2 Qn1 Quebec nonchilled 175 30.4
3 Qn1 Quebec nonchilled 250 34.8> ggplot(CO2) + geom_smooth(aes(x = conc, y = uptake, fill=Treatment))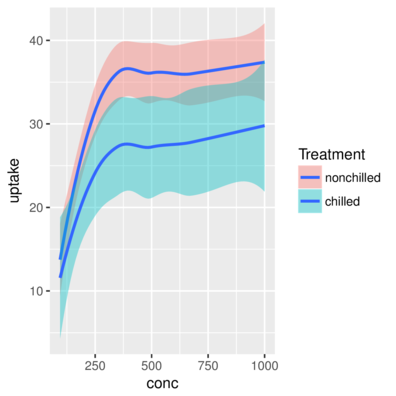
geom_polygon| Feature | geom | stat | position |
|---|---|---|---|
| polygon | _polygon |
_identity |
identity |
> library(maps)
> library(mapdata)
> aus <- map_data("worldHires", region="Australia")
> head(aus,3) long lat group order region subregion
1 142.1461 -10.74943 1 1 Australia Prince of Wales Island
2 142.1430 -10.74525 1 2 Australia Prince of Wales Island
3 142.1406 -10.74113 1 3 Australia Prince of Wales Island> ggplot(aus, aes(x=long, y=lat, group=group)) +
+ geom_polygon()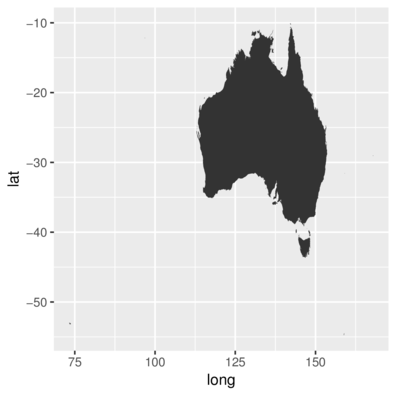
geom_tile| Feature | geom | stat | position |
|---|---|---|---|
| tile | _tile |
_identity |
identity |
> head(faithfuld,3) # A tibble: 3 x 3
eruptions waiting density
<dbl> <dbl> <dbl>
1 1.600000 43 0.003216159
2 1.647297 43 0.003835375
3 1.694595 43 0.004435548> ggplot(faithfuld, aes(waiting, eruptions)) +
+ geom_tile(aes(fill = density))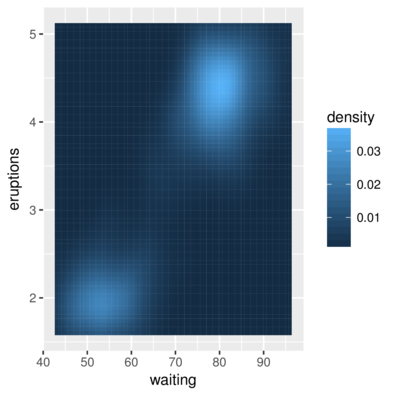
geom_raster| Feature | geom | stat | position |
|---|---|---|---|
| raster | _raster |
_identity |
identity |
> head(faithfuld,3)# A tibble: 3 x 3
eruptions waiting density
<dbl> <dbl> <dbl>
1 1.600000 43 0.003216159
2 1.647297 43 0.003835375
3 1.694595 43 0.004435548> ggplot(faithfuld, aes(waiting, eruptions)) +
+ geom_raster(aes(fill = density))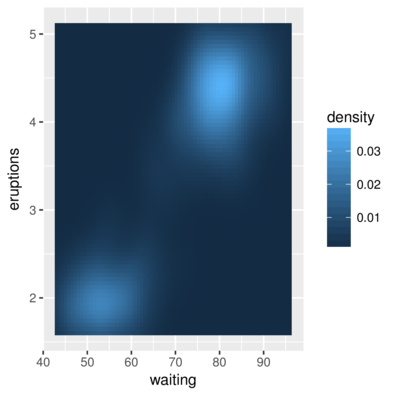
> head(warpbreaks) breaks wool tension
1 26 A L
2 30 A L
3 54 A L
4 25 A L
5 70 A L
6 52 A L> summary(warpbreaks) breaks wool tension
Min. :10.00 A:27 L:18
1st Qu.:18.25 B:27 M:18
Median :26.00 H:18
Mean :28.15
3rd Qu.:34.00
Max. :70.00 geom_errorbar| Feature | geom | stat | position |
|---|---|---|---|
| errorbar | _identity |
_identity |
identity |
> library(dplyr)
> library(gmodels)
> warpbreaks.sum <- warpbreaks %>% group_by(wool) %>%
+ summarise(Mean=mean(breaks), Lower=ci(breaks)[2], Upper=ci(breaks)[3])
> warpbreaks.sum# A tibble: 2 x 4
wool Mean Lower Upper
<fctr> <dbl> <dbl> <dbl>
1 A 31.03704 24.76642 37.30765
2 B 25.25926 21.57994 28.93858geom_errorbar| Feature | geom | stat | position |
|---|---|---|---|
| errorbar | _identity |
_identity |
identity |
> ggplot(warpbreaks.sum) +
+ geom_errorbar(aes(x = wool, ymin = Lower, ymax = Upper))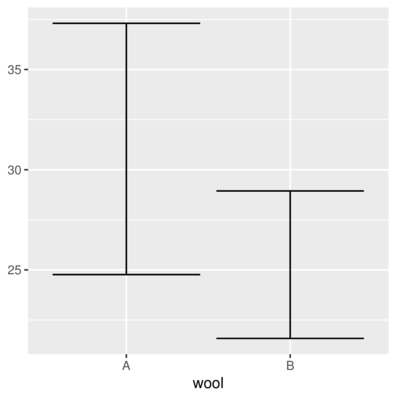
geom_errorbar| Feature | geom | stat | position |
|---|---|---|---|
| errorbar | _identity |
_summary |
identity |
> head(warpbreaks,3) breaks wool tension
1 26 A L
2 30 A L
3 54 A L> ggplot(warpbreaks) + geom_errorbar(aes(x = wool, y = breaks),
+ stat = "summary", fun.data = "mean_cl_boot")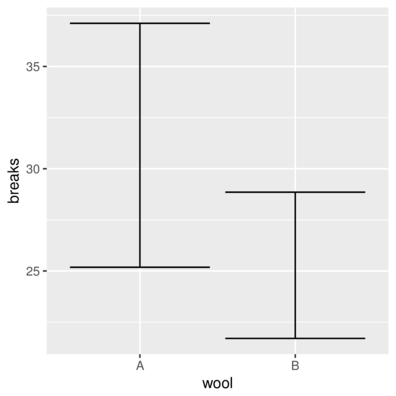
> head(CO2,3) Plant Type Treatment conc uptake
1 Qn1 Quebec nonchilled 95 16.0
2 Qn1 Quebec nonchilled 175 30.4
3 Qn1 Quebec nonchilled 250 34.8> ggplot(CO2)+geom_point(aes(x=conc,y=uptake))+
+ coord_cartesian() #default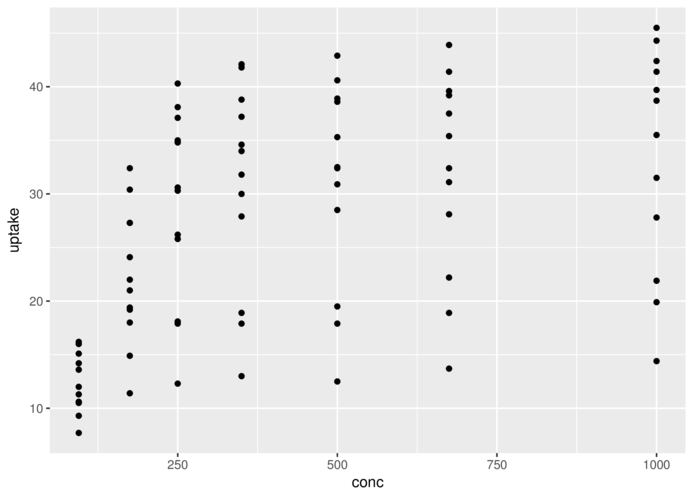
> head(CO2,3) Plant Type Treatment conc uptake
1 Qn1 Quebec nonchilled 95 16.0
2 Qn1 Quebec nonchilled 175 30.4
3 Qn1 Quebec nonchilled 250 34.8> ggplot(CO2)+geom_point(aes(x=conc,y=uptake))+
+ coord_polar()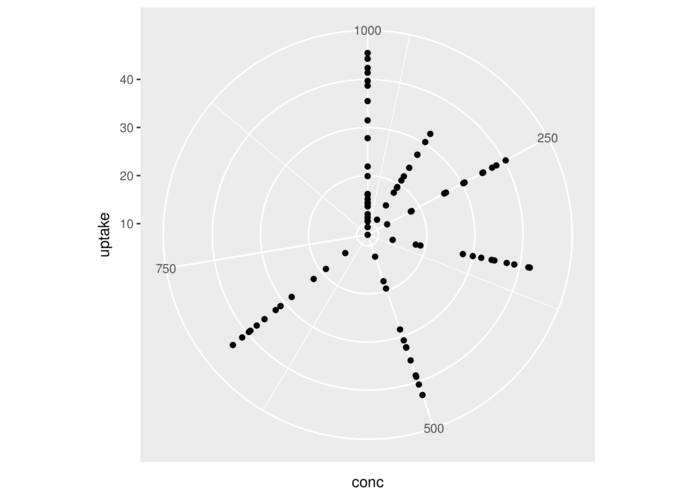
> head(CO2,3) Plant Type Treatment conc uptake
1 Qn1 Quebec nonchilled 95 16.0
2 Qn1 Quebec nonchilled 175 30.4
3 Qn1 Quebec nonchilled 250 34.8> ggplot(CO2)+geom_point(aes(x=conc,y=uptake))+
+ coord_flip()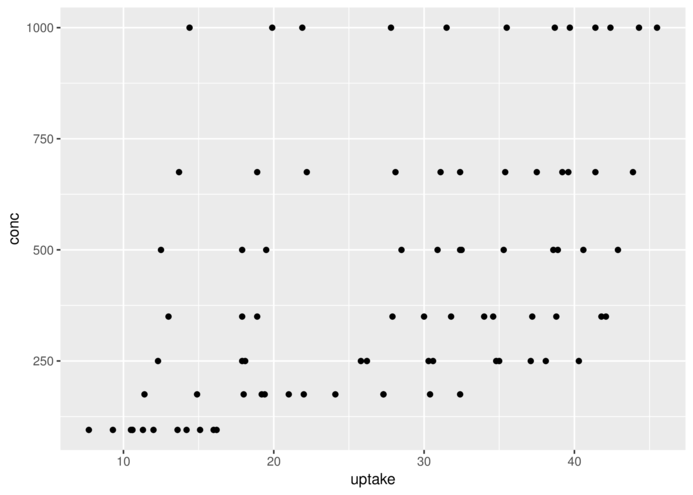
> #Orthographic coordinates
> library(maps)
> library(mapdata)
> aus <- map_data("worldHires", region="Australia")
> ggplot(aus, aes(x=long, y=lat, group=group)) +
+ coord_map("ortho", orientation=c(-20,125,23.5))+
+ geom_polygon()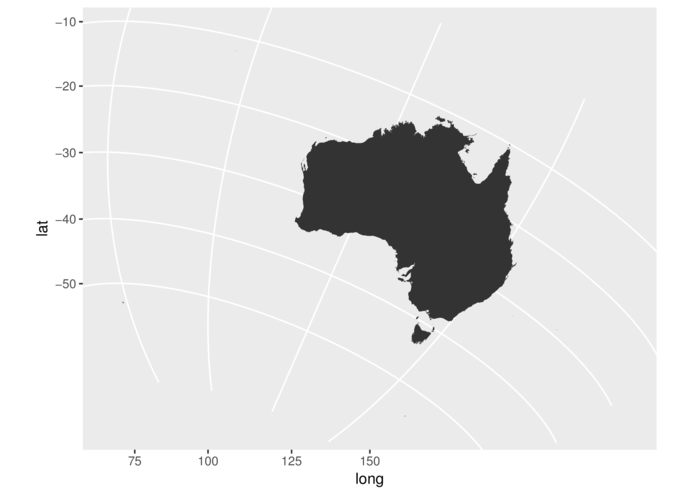
scale_x_ and scale_y_Axis titles
> head(CO2,2) Plant Type Treatment conc uptake
1 Qn1 Quebec nonchilled 95 16.0
2 Qn1 Quebec nonchilled 175 30.4> ggplot(CO2, aes(y=uptake,x=conc)) + geom_point()+
+ scale_x_continuous(name="CO2 conc")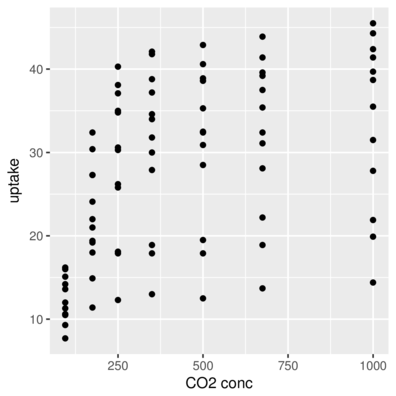
scale_x_ and scale_y_Axis titles with math
> head(CO2,2) Plant Type Treatment conc uptake
1 Qn1 Quebec nonchilled 95 16.0
2 Qn1 Quebec nonchilled 175 30.4> ggplot(CO2, aes(y=uptake,x=conc)) + geom_point()+
+ scale_x_continuous(name=expression(Ambient~CO[2]~concentration~(mg/l)))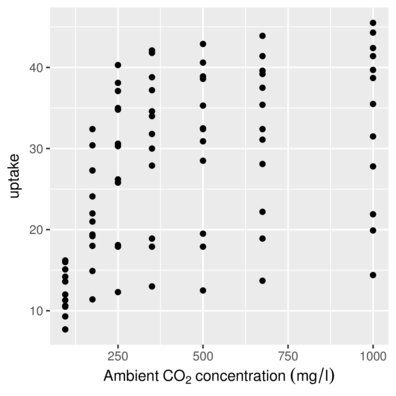
scale_x_ and scale_y_Axis more padding
> ggplot(CO2, aes(y=uptake,x=conc)) + geom_point()+
+ scale_x_continuous(name="CO2 conc", expand=c(0,200))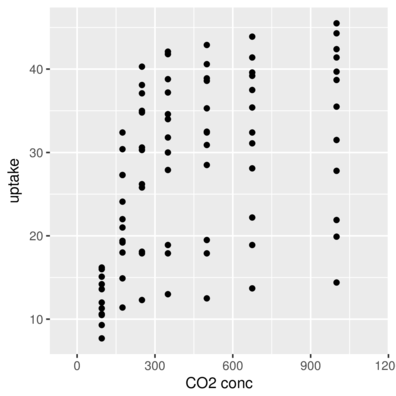
scale_x_ and scale_y_Axis on a log scale
> ggplot(CO2, aes(y=uptake,x=conc)) + geom_point()+
+ scale_x_log10(name="CO2 conc",
+ breaks=as.vector(c(1,2,5,10) %o% 10^(-1:2)))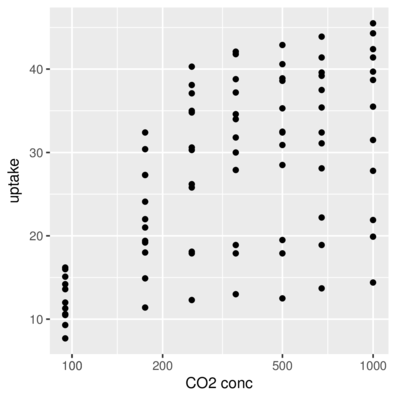
scale_x_ and scale_y_Axis representing categorical data
> ggplot(CO2, aes(y=uptake,x=Treatment)) + geom_point()+
+ scale_x_discrete(name="Treatment")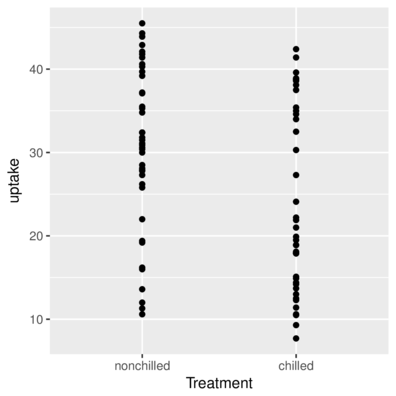
scale_sizeSize according to continuous variable
> state=data.frame(state.x77,state.region, state.division,state.center) %>%
+ select(Illiteracy,state.region,x,y)
> head(state,2) Illiteracy state.region x y
Alabama 2.1 South -86.7509 32.5901
Alaska 1.5 West -127.2500 49.2500> ggplot(state, aes(y=y,x=x)) + geom_point(aes(size=Illiteracy))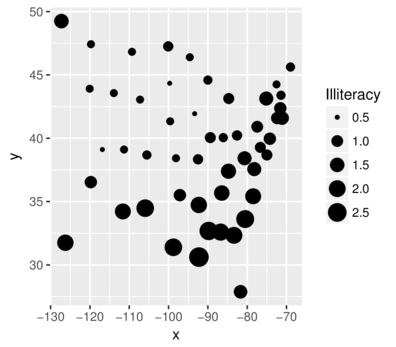
scale_sizeDiscrete sizes ranging in size from 2 to 4
> head(state,2) Illiteracy state.region x y
Alabama 2.1 South -86.7509 32.5901
Alaska 1.5 West -127.2500 49.2500> ggplot(state, aes(y=y,x=x)) + geom_point(aes(size=state.region))+
+ scale_size_discrete(name="Region", range=c(2,10))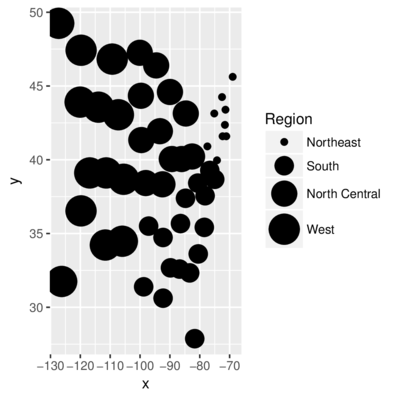
scale_sizeManual sizes (2 and 4)
> head(state,2) Illiteracy state.region x y
Alabama 2.1 South -86.7509 32.5901
Alaska 1.5 West -127.2500 49.2500> ggplot(state, aes(y=y,x=x)) + geom_point(aes(size=state.region))+
+ scale_size_manual(name="Region", values=c(2,5,6,10))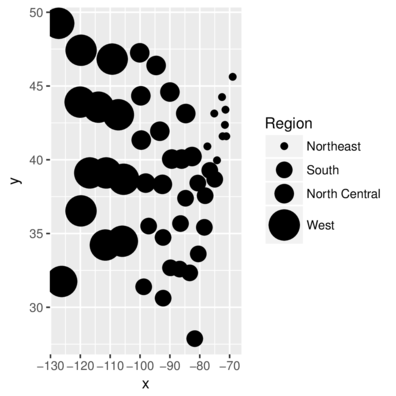
scale_shape> head(CO2,2) Plant Type Treatment conc uptake
1 Qn1 Quebec nonchilled 95 16.0
2 Qn1 Quebec nonchilled 175 30.4> ggplot(CO2, aes(y=uptake,x=conc)) + geom_point(aes(shape=Treatment))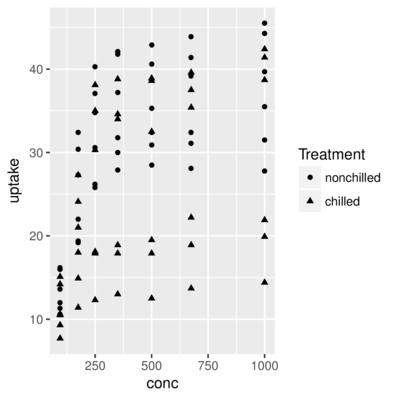
scale_shape> CO2 = CO2 %>% mutate(Comb=interaction(Type, Treatment))
> ggplot(CO2, aes(y=uptake,x=conc)) + geom_point(aes(shape=Comb))+
+ scale_shape_discrete(name="Type",
+ breaks=c("Quebec.nonchilled","Quebec.chilled",
+ "Mississippi.nonchilled","Mississippi.chilled"),
+ labels=c("Quebec non-chilled","Quebec chilled",
+ "Miss. non-chilled","Miss. chilled"))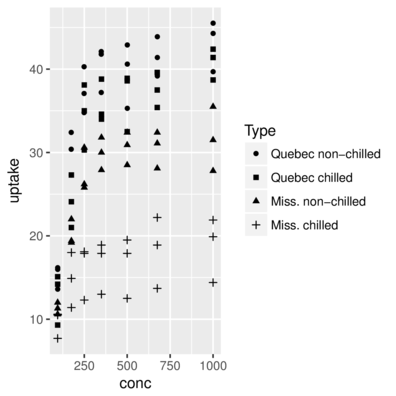
scale_linetype> head(CO2,2) Plant Type Treatment conc uptake Comb
1 Qn1 Quebec nonchilled 95 16.0 Quebec.nonchilled
2 Qn1 Quebec nonchilled 175 30.4 Quebec.nonchilled> ggplot(CO2, aes(y=uptake,x=conc)) + geom_smooth(aes(linetype=Comb))+
+ scale_linetype_discrete(name="Type")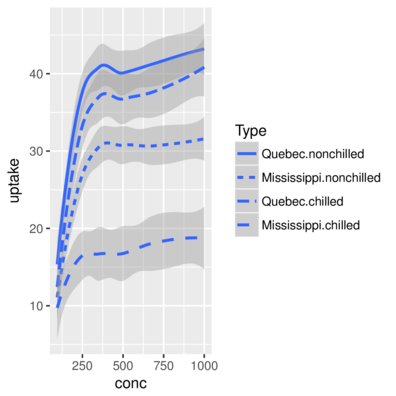
scale_linetype> head(CO2,2) Plant Type Treatment conc uptake Comb
1 Qn1 Quebec nonchilled 95 16.0 Quebec.nonchilled
2 Qn1 Quebec nonchilled 175 30.4 Quebec.nonchilled> ggplot(CO2, aes(y=uptake,x=conc)) + geom_smooth(aes(linetype=Treatment))+
+ scale_linetype_manual(name="Treatment", values=c("dashed","dotted"))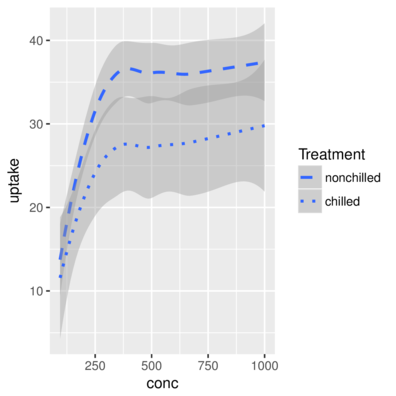
scale_fill and scale_color> head(faithfuld,2)# A tibble: 2 x 3
eruptions waiting density
<dbl> <dbl> <dbl>
1 1.600000 43 0.003216159
2 1.647297 43 0.003835375> ggplot(faithfuld, aes(waiting, eruptions)) +
+ geom_raster(aes(fill = density)) +
+ scale_fill_continuous(low='red',high='blue')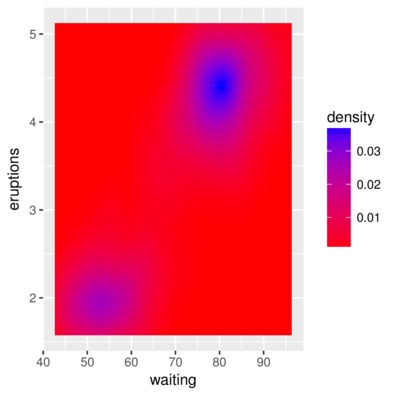
Panels - matrices of plots
facet_wrapfacet_grid> ggplot(CO2)+geom_point(aes(x=conc,y=uptake, colour=Type))+
+ facet_wrap(~Plant)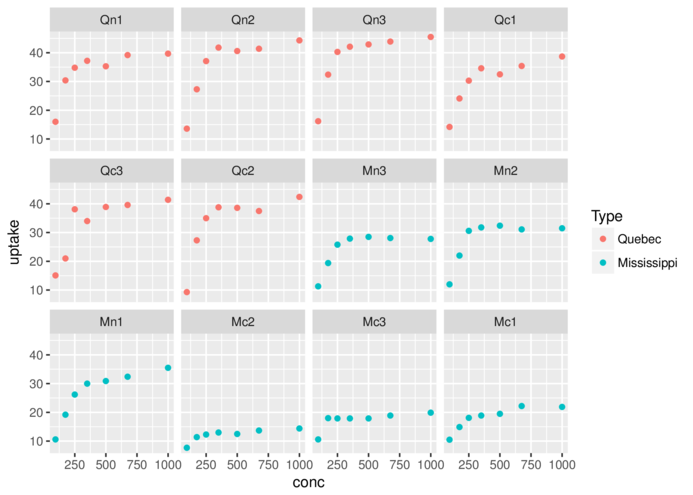
> ggplot(CO2)+geom_line(aes(x=conc,y=uptake, colour=Type))+
+ facet_wrap(~Plant, scales='free_y')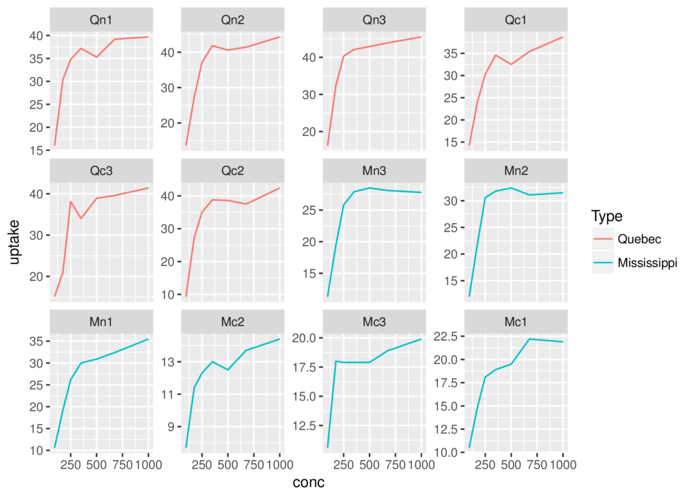
> ggplot(CO2)+geom_point(aes(x=conc,y=uptake, colour=Type))+
+ facet_grid(Type~Treatment)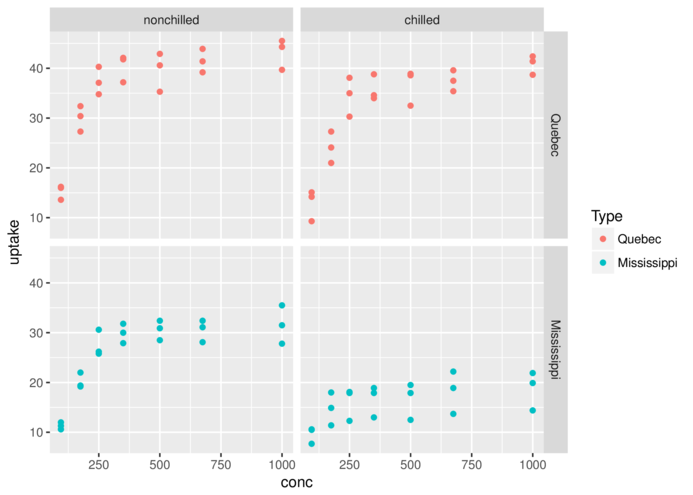
theme_classic> ggplot(CO2, aes(y = uptake, x = conc)) + geom_smooth() +
+ geom_point() + theme_classic()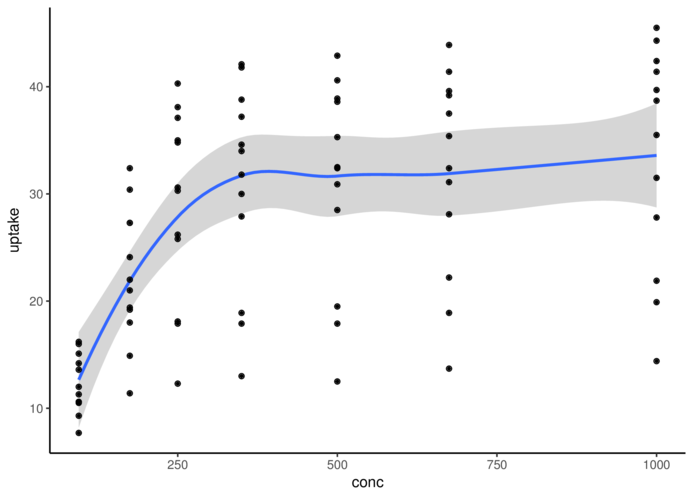
theme_bw> ggplot(CO2, aes(y = uptake, x = conc)) + geom_smooth() +
+ geom_point() + theme_bw()
theme_grey> ggplot(CO2, aes(y = uptake, x = conc)) + geom_smooth() +
+ geom_point() + theme_grey()
theme_minimal> ggplot(CO2, aes(y = uptake, x = conc)) + geom_smooth() +
+ geom_point() + theme_minimal()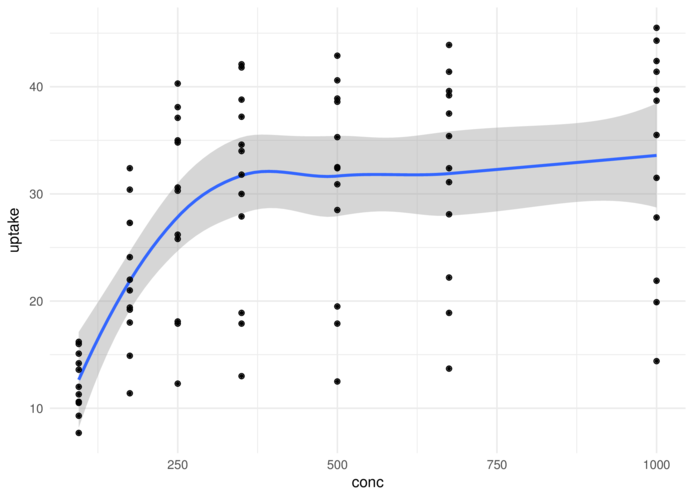
theme_linedraw> ggplot(CO2, aes(y = uptake, x = conc)) + geom_smooth() +
+ geom_point() + theme_linedraw()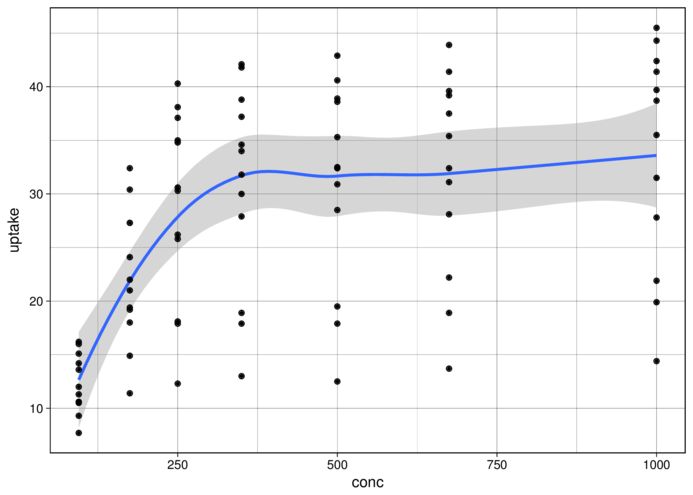
theme_light> ggplot(CO2, aes(y = uptake, x = conc)) + geom_smooth() +
+ geom_point() + theme_light()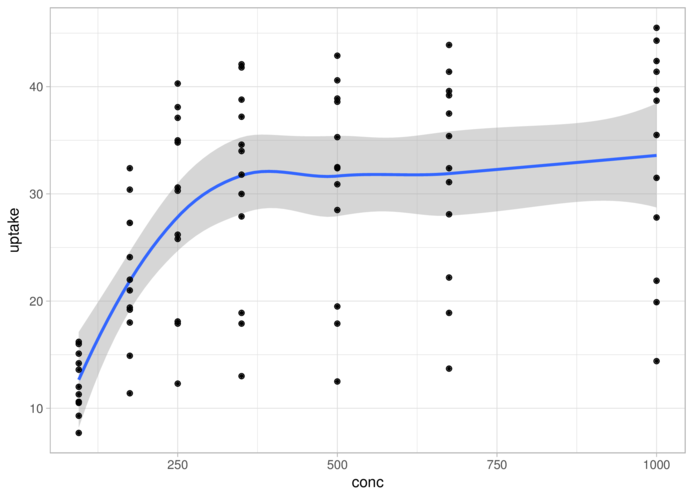
> png('images/xkcd.png', width=500, height=500, res=200)
> library(xkcd)
> library(sysfonts)
> #library(extrafont)
> #download.file("http://simonsoftware.se/other/xkcd.ttf", dest="xkcd.ttf")
> ##font_import(".")
> #loadfonts()
> xrange <- range(CO2$conc)
> yrange <- range(CO2$uptake)
> ggplot(CO2, aes(y = uptake, x = conc)) + geom_smooth(position='jitter', size=1.5) +
+ #geom_point() +
+ theme_minimal()+theme(text=element_text(size=16, family='xkcd'))+
+ xkcdaxis(xrange, yrange)
>
> dev.off() 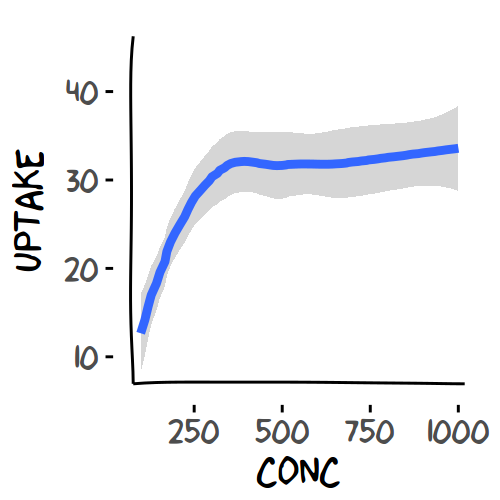
> head(state) Illiteracy state.region x y
Alabama 2.1 South -86.7509 32.5901
Alaska 1.5 West -127.2500 49.2500
Arizona 1.8 West -111.6250 34.2192
Arkansas 1.9 South -92.2992 34.7336
California 1.1 West -119.7730 36.5341
Colorado 0.7 West -105.5130 38.6777Calculate the mean and 95% confidence interval of Illiteracy per state.region and plot them. and plot them
> head(state) Illiteracy state.region x y
Alabama 2.1 South -86.7509 32.5901
Alaska 1.5 West -127.2500 49.2500
Arizona 1.8 West -111.6250 34.2192
Arkansas 1.9 South -92.2992 34.7336
California 1.1 West -119.7730 36.5341
Colorado 0.7 West -105.5130 38.6777> library(gmodels)
> state.sum = state %>% group_by(state.region) %>%
+ summarise(Mean=mean(Illiteracy), Lower=ci(Illiteracy)[2],
+ Upper=ci(Illiteracy)[3])
> state.sum# A tibble: 4 x 4
state.region Mean Lower Upper
<fctr> <dbl> <dbl> <dbl>
1 Northeast 1.000000 0.7860119 1.2139881
2 South 1.737500 1.4431367 2.0318633
3 North Central 0.700000 0.6101452 0.7898548
4 West 1.023077 0.6553719 1.3907819> ggplot(state.sum, aes(y=Mean, x=state.region)) + geom_point() +
+ geom_errorbar(aes(ymin=Lower, ymax=Upper), width=0.1)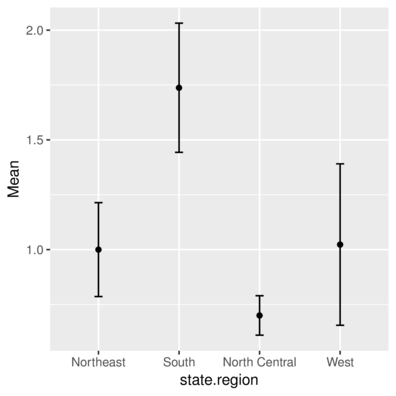
> ggplot(state.sum, aes(y=Mean, x=state.region)) + geom_point() +
+ geom_errorbar(aes(ymin=Lower, ymax=Upper), width=0.1) +
+ scale_x_discrete('Region') +
+ scale_y_continuous('Illiteracy rate (%)')+
+ theme_classic() +
+ theme(axis.line.y=element_line(),axis.line.x=element_line())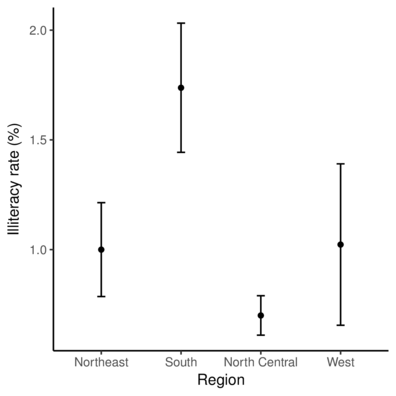
Overlay illiteracy data onto map of US
> library(mapdata)
> US <- map_data("worldHires", region="USA")
> ggplot(US) +
+ geom_polygon(aes(x=long, y=lat, group=group)) +
+ geom_point(data=state,aes(y=y,x=x, size=Illiteracy),color='red')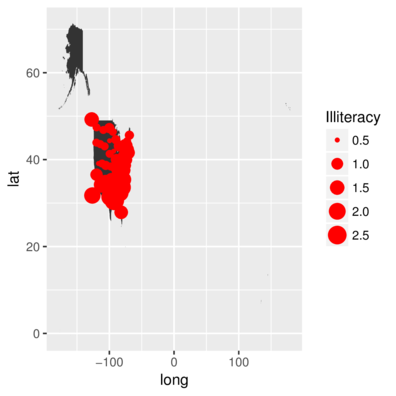
Overlay illiteracy data onto map of US
> library(mapdata)
> US <- map_data("worldHires", region="USA")
> ggplot(US) +
+ geom_polygon(aes(x=long, y=lat, group=group)) +
+ geom_point(data=state,aes(y=y,x=x, size=Illiteracy),color='red')+
+ coord_map(xlim=c(-150,-50),ylim=c(20,60)) + theme_minimal()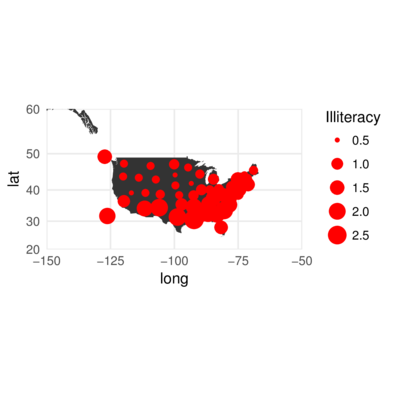
> MACNALLY <- read.csv('../data/macnally.csv',
+ header=T, row.names=1, strip.white=TRUE)
> head(MACNALLY) HABITAT GST EYR
Reedy Lake Mixed 3.4 0.0
Pearcedale Gipps.Manna 3.4 9.2
Warneet Gipps.Manna 8.4 3.8
Cranbourne Gipps.Manna 3.0 5.0
Lysterfield Mixed 5.6 5.6
Red Hill Mixed 8.1 4.1Calculate the mean and standard error of GST and plot them
Calculate the mean and standard error of GST and plot mean and confidence bars
> library(gmodels)
> ci(MACNALLY$GST) Estimate CI lower CI upper Std. Error
4.878378 4.035292 5.721465 0.415704 > MACNALLY.agg = MACNALLY %>% group_by(HABITAT) %>%
+ summarize(Mean=mean(GST), Lower=ci(GST)[2], Upper=ci(GST)[3])
> ggplot(MACNALLY.agg, aes(y=Mean, x=HABITAT)) +
+ geom_errorbar(aes(ymin=Lower, ymax=Upper), width=0.1)+
+ geom_point() + theme_classic()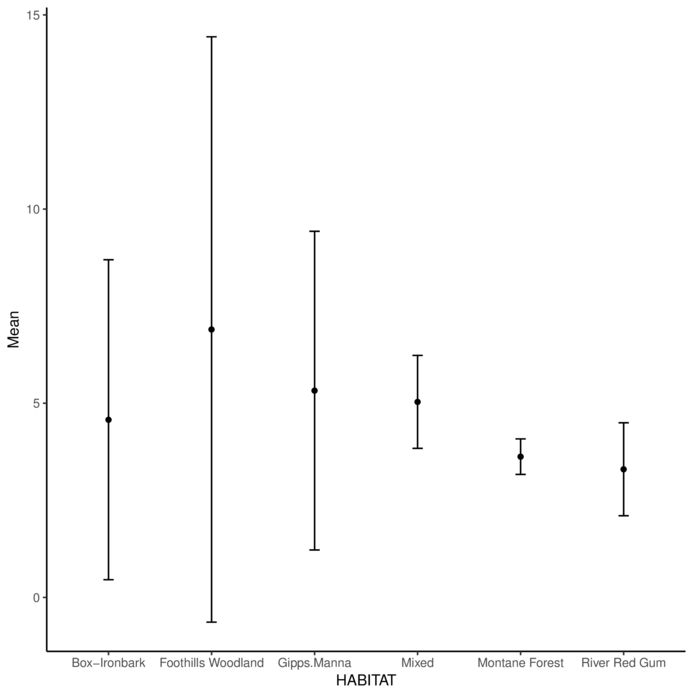
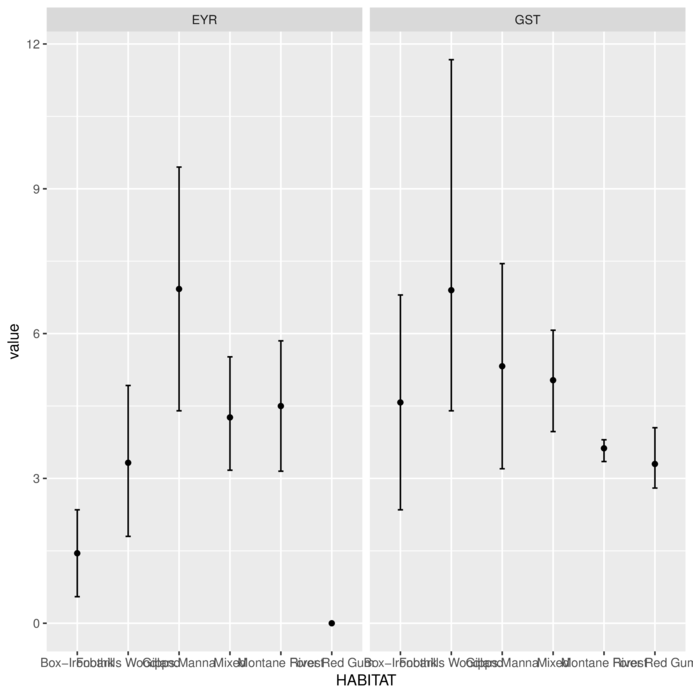
You can also use ggplot’s summary
> library(tidyverse)
> MACNALLY.melt = MACNALLY %>% gather(key=variable, value=value,-HABITAT)
> ggplot(MACNALLY.melt, aes(y=value, x=HABITAT)) +
+ stat_summary(fun.y='mean', geom='point')+
+ stat_summary(fun.data='mean_cl_normal', geom='errorbar', width=0.1)+
+ facet_grid(~variable)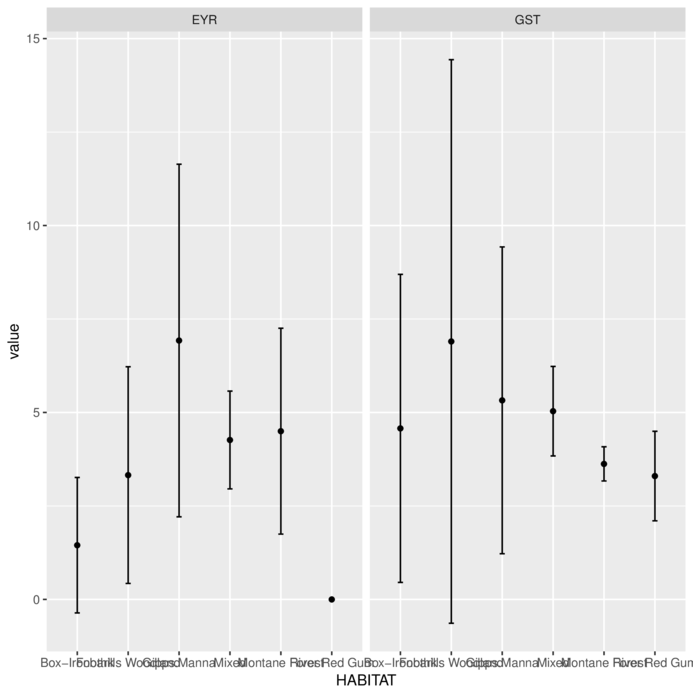
> #and bootstrapped means..
> ggplot(MACNALLY.melt, aes(y=value, x=HABITAT)) +
+ stat_summary(fun.y='mean', geom='point')+
+ stat_summary(fun.data='mean_cl_boot', geom='errorbar', width=0.1)+
+ facet_grid(~variable)