Mathematical models
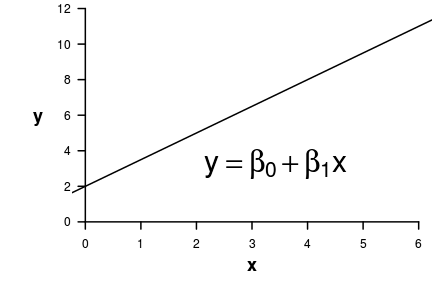
Use samples to:
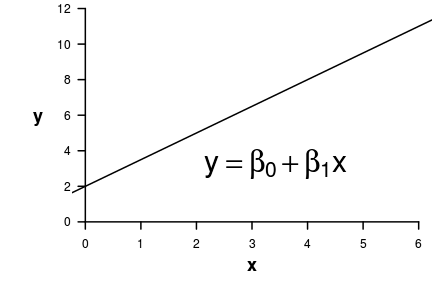
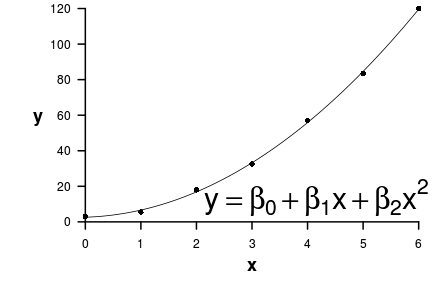
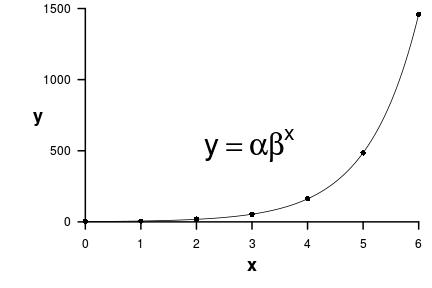
\[
\begin{aligned}
y_i &= \qquad\beta_0\qquad + \qquad\beta_1\quad\times \quad x_1 \qquad+ \qquad\epsilon_1\\[1em]
\begin{matrix}response\\variable\end{matrix}&=
\underbrace{
\underbrace{\begin{matrix}population\\intercept\end{matrix}}_\text{intercept term}
\quad +
\underbrace{\begin{matrix}population\\slope\end{matrix}
\times
\begin{matrix}predictor\\variable\end{matrix}}_\text{slope term}
}_\text{Systematic component}
+
\underbrace{error}_\text{Stoichastic component}
\end{aligned}
\]
\[
\begin{aligned}
y_i &= \qquad\beta_0\qquad + \qquad\beta_1\quad\times \quad x_1 \qquad+ \qquad\varepsilon_1\\[1em]
\begin{matrix}response\\\mathbf{vector}\end{matrix}&=
\underbrace{
\underbrace{\begin{matrix}intercept\\\mathbf{single~value}\end{matrix}}_\text{intercept term}
\quad +
\underbrace{\begin{matrix}slope\\\mathbf{single~value}\end{matrix}
\times
\begin{matrix}predictor\\\mathbf{vector}\end{matrix}}_\text{slope term}
}_\text{Systematic component}
+
\underbrace{error}_{\text{Stoichastic component}}
\end{aligned}
\]
| Vector | Matrix |
|---|---|
| \(\begin{pmatrix}3.0\\2.5\\6.0\\5.5\\9.0\\8.6\\12.0\end{pmatrix}\) | \(\begin{pmatrix}1&0\\1&1\\1&2\\1&3\\1&4\\1&5\\1&6\end{pmatrix}\) |
| Has length ONLY | Has length AND width |

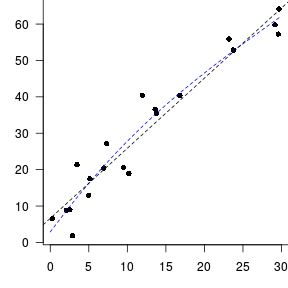
Ordinary Least Squares
| Y | X |
|---|---|
| 3 | 0 |
| 2.5 | 1 |
| 6 | 2 |
| 5.5 | 3 |
| 9 | 4 |
| 8.6 | 5 |
| 12 | 6 |
Ho:\(\beta_1=0\) (slope equals zero)
The t-statistic
\[t=\frac{param}{SE_{param}}\] \[t=\frac{\beta_1}{SE_{\beta_1}}\]
Ho:\(\beta_1=0\) (slope equals zero)
The t-statistic and the t distribution
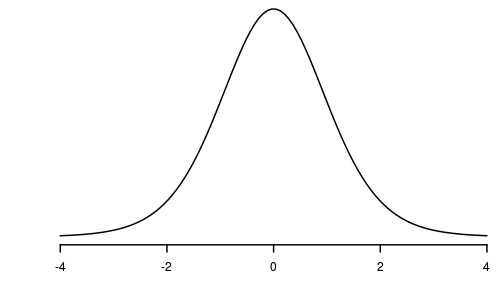




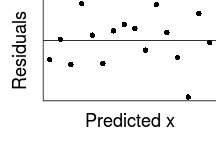


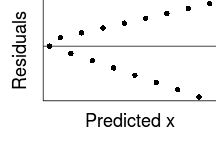
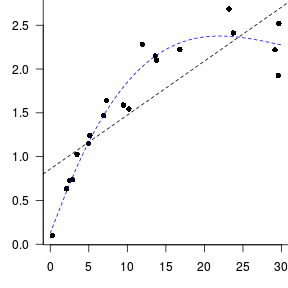
Trendline
Loess (lowess) smoother
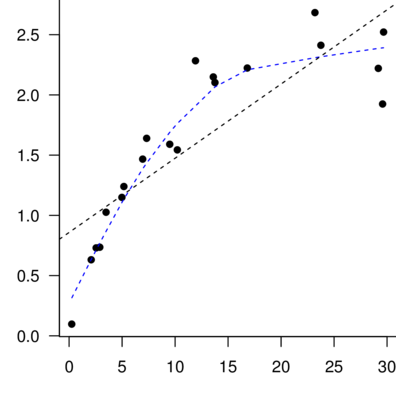
Spline smoother
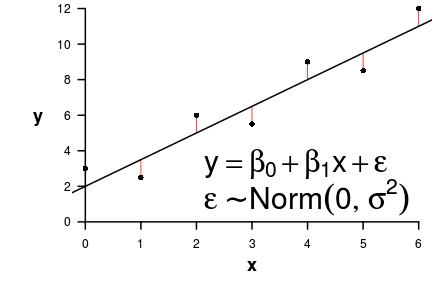
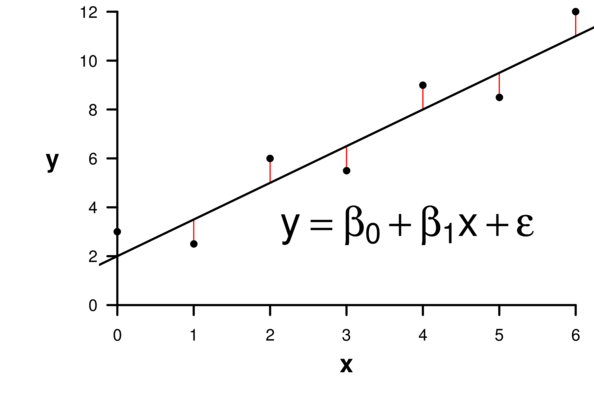
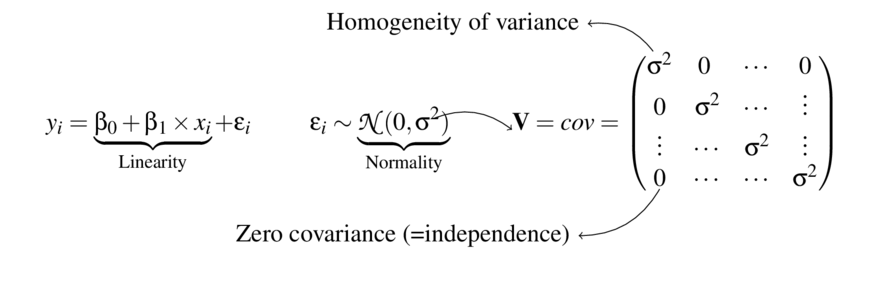
\[ \begin{align*} y_{i} &= \beta_0 + \beta_1 \times x_{i} + \varepsilon_i\\ \epsilon_i&\sim\mathcal{N}(0, \sigma^2) \\ \end{align*} \]
\[ \begin{align*} y_{i} &= \beta_0 + \beta_1 \times x_{i} + \varepsilon_i\\ \epsilon_i&\sim\mathcal{N}(0, \sigma^2) \\ \end{align*} \]
Make these data and call the data frame DATA
| Y | X |
|---|---|
| 3 | 0 |
| 2.5 | 1 |
| 6 | 2 |
| 5.5 | 3 |
| 9 | 4 |
| 8.6 | 5 |
| 12 | 6 |
Make these data and call the data frame DATA
| Y | X |
|---|---|
| 3 | 0 |
| 2.5 | 1 |
| 6 | 2 |
| 5.5 | 3 |
| 9 | 4 |
| 8.6 | 5 |
| 12 | 6 |
> DATA <- data.frame(Y=c(3, 2.5, 6.0, 5.5, 9.0, 8.6, 12), X=0:6)| Format of fertilizer.csv data files | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|

|
||||||||||||||||||
> fert <- read.csv('../data/fertilizer.csv', strip.white=T)
> fert FERTILIZER YIELD
1 25 84
2 50 80
3 75 90
4 100 154
5 125 148
6 150 169
7 175 206
8 200 244
9 225 212
10 250 248> head(fert) FERTILIZER YIELD
1 25 84
2 50 80
3 75 90
4 100 154
5 125 148
6 150 169> summary(fert) FERTILIZER YIELD
Min. : 25.00 Min. : 80.0
1st Qu.: 81.25 1st Qu.:104.5
Median :137.50 Median :161.5
Mean :137.50 Mean :163.5
3rd Qu.:193.75 3rd Qu.:210.5
Max. :250.00 Max. :248.0 > str(fert)'data.frame': 10 obs. of 2 variables:
$ FERTILIZER: int 25 50 75 100 125 150 175 200 225 250
$ YIELD : int 84 80 90 154 148 169 206 244 212 248> library(INLA)
>
> fert.inla <- inla(YIELD ~ FERTILIZER, data=fert)
> summary(fert.inla)Call: “inla(formula = YIELD ~ FERTILIZER, data = fert)”
Time used: Pre-processing Running inla Post-processing Total 0.3043 0.0715 0.0217 0.3974
Fixed effects: mean sd 0.025quant 0.5quant 0.975quant mode kld (Intercept) 51.9341 12.9747 25.9582 51.9335 77.8990 51.9339 0 FERTILIZER 0.8114 0.0836 0.6439 0.8114 0.9788 0.8114 0
The model has no random effects
Model hyperparameters: mean sd 0.025quant 0.5quant 0.975quant mode Precision for the Gaussian observations 0.0035 0.0015 0.0012 0.0032 0.007 0.0028
Expected number of effective parameters(std dev): 2.00(0.00) Number of equivalent replicates : 5.00
Marginal log-Likelihood: -61.65
Question: is there a relationship between fertilizer concentration and grass yeild?
Linear model:> library(car)
> scatterplot(Y~X, data=DATA)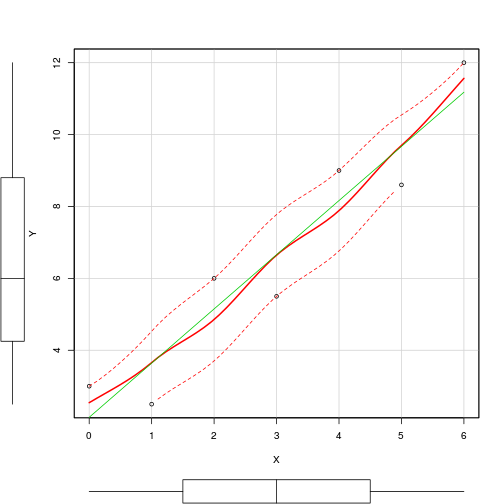
> library(car)
> peake <- read.csv('../data/peake.csv')
> scatterplot(SPECIES ~ AREA, data=peake)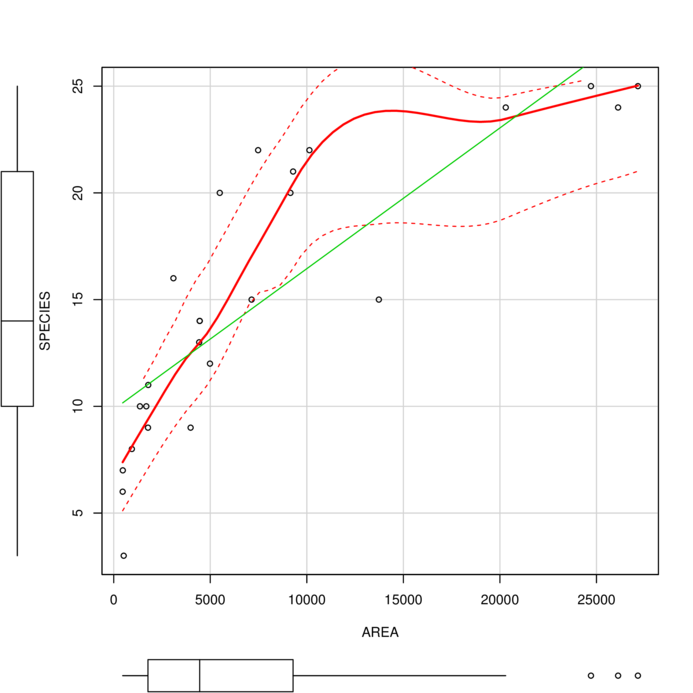
> scatterplot(SPECIES ~ AREA, data=peake,
+ smoother=gamLine)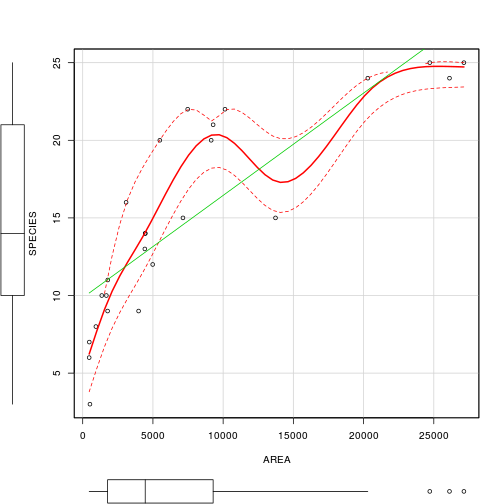
> library(ggplot2)
> library(gridExtra)
> ggplot(peake, aes(y=SPECIES, x=AREA)) + geom_point()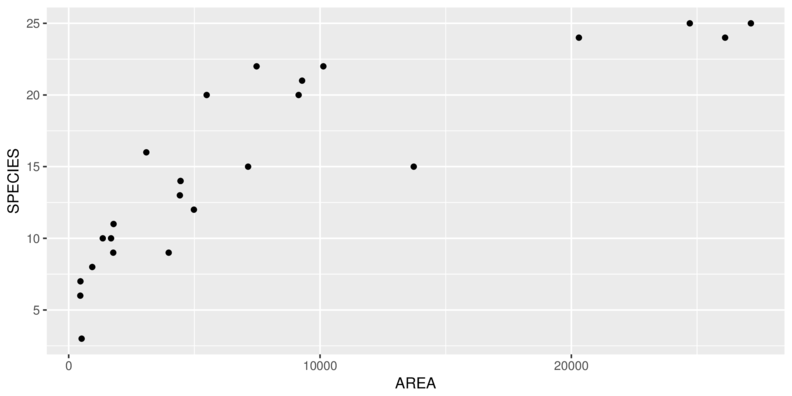
> ggplot(peake, aes(y=SPECIES, x=AREA)) + geom_point() +
+ geom_smooth()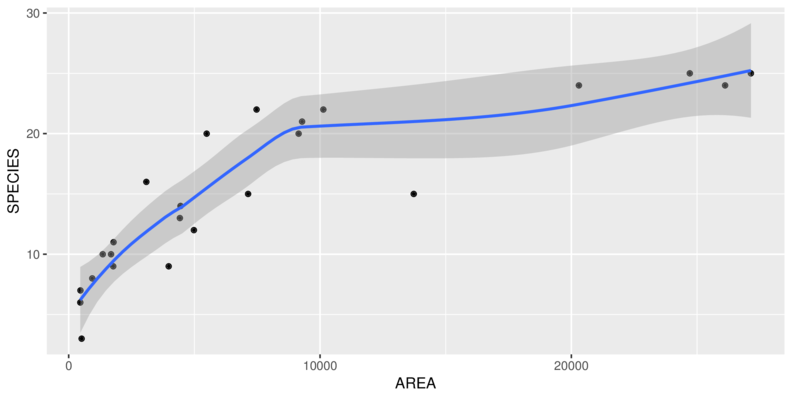
> p2 <- ggplot(peake, aes(y=SPECIES, x=1)) + geom_boxplot()
> p3 <- ggplot(peake, aes(y=AREA, x=1)) + geom_boxplot()
> grid.arrange(p1,p2,p3, ncol=3)Error in arrangeGrob(...): object 'p1' not found> lm(formula, data= DATAFRAME)| Model | R formula | Description |
|---|---|---|
| \(y_i = \beta_0 + \beta_1 x_i\) | y~1+x |
|
y~x |
Full model | |
| \(y_i = \beta_0\) | y~1 |
Null model |
| \(y_i = \beta_1\) | y~-1+x |
Through origin |
\[y_i = \beta_0 + \beta_1 x_i \qquad N(0,\sigma)\]
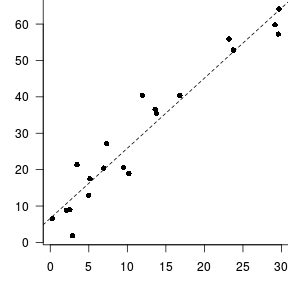
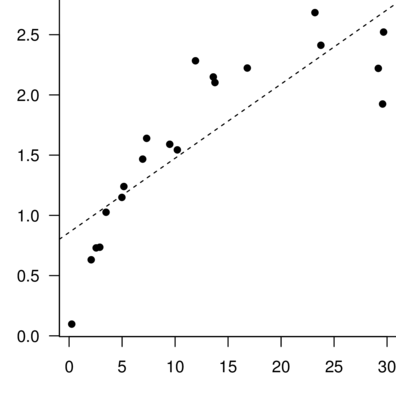
> DATA.lm<-lm(Y~X, data=DATA)
TIME TO FIT A MODEL

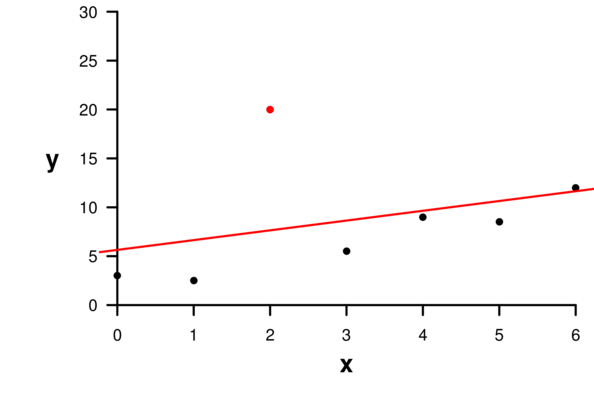
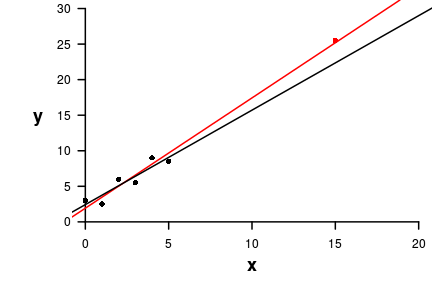
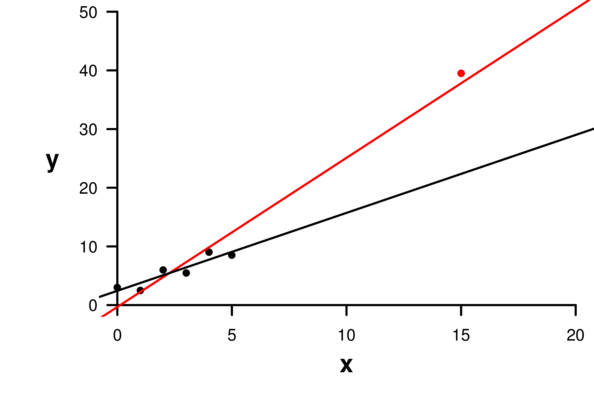
| Extractor | Description |
|---|---|
residuals() |
Extracts residuals from model |
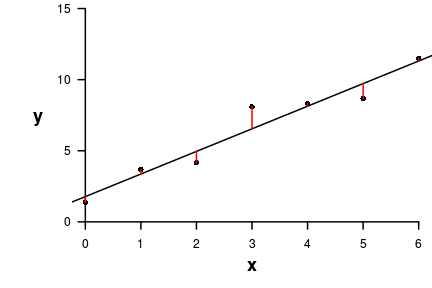
> residuals(DATA.lm) 1 2 3 4 5 6 7
0.8642857 -1.1428571 0.8500000 -1.1571429 0.8357143 -1.0714286 0.8214286 | Extractor | Description |
|---|---|
residuals() |
Extracts residuals from model |
fitted() |
Extracts the predicted values |
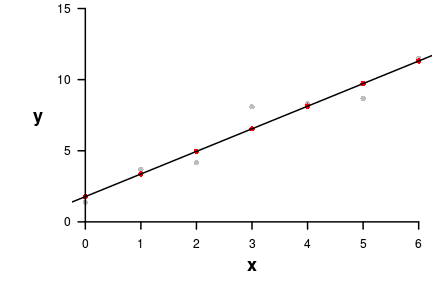
> fitted(DATA.lm) 1 2 3 4 5 6 7
2.135714 3.642857 5.150000 6.657143 8.164286 9.671429 11.178571 | Extractor | Description |
|---|---|
residuals() |
Extracts residuals from model |
fitted() |
Extracts the predicted values |
plot() |
Series of diagnostic plots |
> plot(DATA.lm)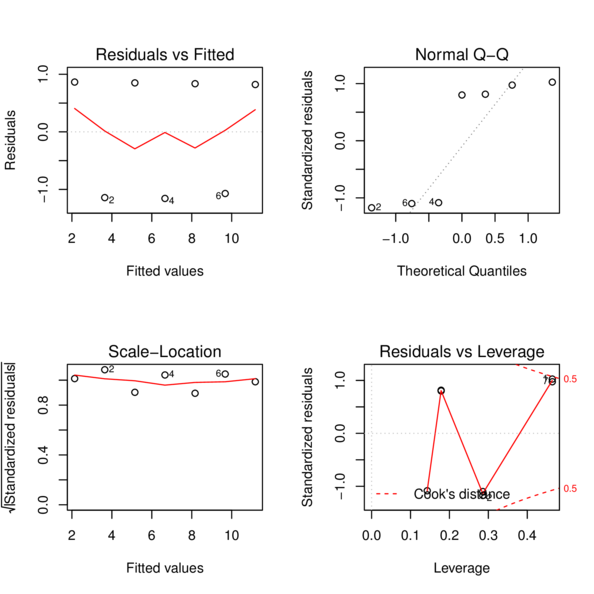
| Extractor | Description |
|---|---|
residuals() |
Residuals |
fitted() |
Predicted values |
plot() |
Diagnostic plots |
influence.measures() |
Leverage (hat) and Cook’s D |
> influence.measures(DATA.lm)Influence measures of
lm(formula = Y ~ X, data = DATA) :
dfb.1_ dfb.X dffit cov.r cook.d hat inf
1 0.9603 -7.99e-01 0.960 1.82 0.4553 0.464
2 -0.7650 5.52e-01 -0.780 1.15 0.2756 0.286
3 0.3165 -1.63e-01 0.365 1.43 0.0720 0.179
4 -0.2513 -7.39e-17 -0.453 1.07 0.0981 0.143
5 0.0443 1.60e-01 0.357 1.45 0.0696 0.179
6 0.1402 -5.06e-01 -0.715 1.26 0.2422 0.286
7 -0.3466 7.50e-01 0.901 1.91 0.4113 0.464 | Extractor | Description |
|---|---|
residuals() |
Residuals |
fitted() |
Predicted values |
plot() |
Diagnostic plots |
influence.measures() |
Leverage, Cook’s D |
summary() |
Summarizes important output from model |
> summary(DATA.lm)
Call:
lm(formula = Y ~ X, data = DATA)
Residuals:
1 2 3 4 5 6 7
0.8643 -1.1429 0.8500 -1.1571 0.8357 -1.0714 0.8214
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 2.1357 0.7850 2.721 0.041737 *
X 1.5071 0.2177 6.923 0.000965 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.152 on 5 degrees of freedom
Multiple R-squared: 0.9055, Adjusted R-squared: 0.8866
F-statistic: 47.92 on 1 and 5 DF, p-value: 0.0009648| Extractor | Description |
|---|---|
residuals() |
Residuals |
fitted() |
Predicted values |
plot() |
Diagnostic plots |
influence.measures() |
Leverage, Cook’s D |
summary() |
Model output |
confint() |
Confidence intervals of parameters |
> confint(DATA.lm) 2.5 % 97.5 %
(Intercept) 0.1178919 4.153537
X 0.9474996 2.066786| Extractor | Description |
|---|---|
residuals() |
Residuals |
fitted() |
Predicted values |
plot() |
Diagnostic plots |
influence.measures() |
Leverage, Cook’s D |
summary() |
Model output |
confint() |
Confidence intervals |
predict() |
Predict responses to new levels of predictors |
> predict(DATA.lm, newdata=data.frame(X=c(2.5, 4.1)),
+ se=TRUE)$fit
1 2
5.903571 8.315000
$se.fit
1 2
0.4488222 0.4969340
$df
[1] 5
$residual.scale
[1] 1.152017> predict(DATA.lm, newdata=data.frame(X=c(2.5, 4.1)),
+ interval='confidence') fit lwr upr
1 5.903571 4.749837 7.057306
2 8.315000 7.037591 9.592409> predict(DATA.lm, newdata=data.frame(X=c(2.5, 4.1)),
+ interval='prediction') fit lwr upr
1 5.903571 2.725409 9.081734
2 8.315000 5.089881 11.540119| Extractor | Description |
|---|---|
residuals() |
Residuals |
fitted() |
Predicted values |
plot() |
Diagnostic plots |
influence.measures() |
Leverage, Cook’s D |
summary() |
Model output |
confint() |
Confidence intervals |
predict() |
Predict new responses |
plot(allEffects()) |
Effects plots |
> library(effects)
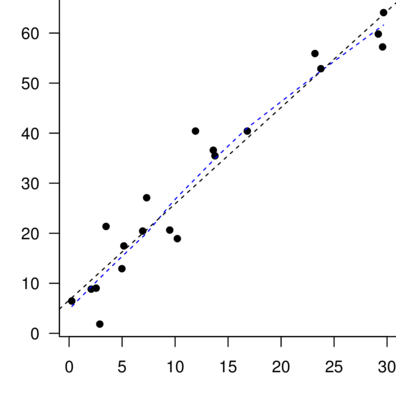
> plot(allEffects(DATA.lm))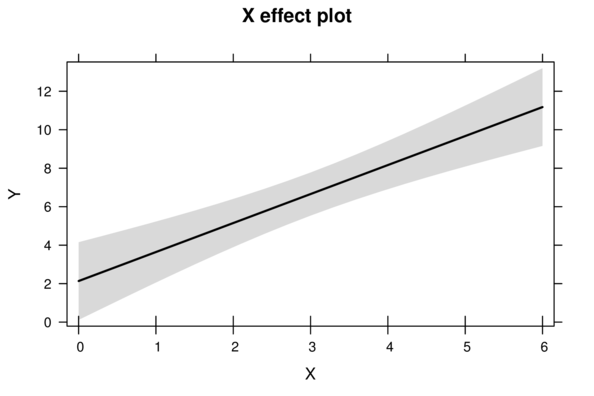
| Format of fertilizer.csv data files | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|

|
||||||||||||||||||
> fert <- read.csv('../data/fertilizer.csv', strip.white=T)
> fert FERTILIZER YIELD
1 25 84
2 50 80
3 75 90
4 100 154
5 125 148
6 150 169
7 175 206
8 200 244
9 225 212
10 250 248> head(fert) FERTILIZER YIELD
1 25 84
2 50 80
3 75 90
4 100 154
5 125 148
6 150 169> summary(fert) FERTILIZER YIELD
Min. : 25.00 Min. : 80.0
1st Qu.: 81.25 1st Qu.:104.5
Median :137.50 Median :161.5
Mean :137.50 Mean :163.5
3rd Qu.:193.75 3rd Qu.:210.5
Max. :250.00 Max. :248.0 > str(fert)'data.frame': 10 obs. of 2 variables:
$ FERTILIZER: int 25 50 75 100 125 150 175 200 225 250
$ YIELD : int 84 80 90 154 148 169 206 244 212 248Question: is there a relationship between fertilizer concentration and grass yeild?
Linear model:| Format of peakquinn.csv data files | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|

|
||||||||||||||||||
> peake <- read.csv('../data/peakquinn.csv', strip.white=T)
> head(peake) AREA INDIV
1 516.00 18
2 469.06 60
3 462.25 57
4 938.60 100
5 1357.15 48
6 1773.66 118> summary(peake) AREA INDIV
Min. : 462.2 Min. : 18.0
1st Qu.: 1773.7 1st Qu.: 148.0
Median : 4451.7 Median : 338.0
Mean : 7802.0 Mean : 446.9
3rd Qu.: 9287.7 3rd Qu.: 632.0
Max. :27144.0 Max. :1402.0 Question: is there a relationship between mussel clump area and number of individuals?
Linear model: