Multiple Linear Regression
Additive model
\(growth = intercept + temperature + nitrogen\)
\(y_i=\beta_0+\beta_1x_{i1}+\beta_2x_{i2}+...+\beta_jx_{ij}+\epsilon_i\)
| Y | X1 | X2 |
|---|---|---|
| 3 | 22.7 | 0.9 |
| 2.5 | 23.7 | 0.5 |
| 6 | 25.7 | 0.6 |
| 5.5 | 29.1 | 0.7 |
| 9 | 22 | 0.8 |
| 8.6 | 29 | 1.3 |
| 12 | 29.4 | 1 |
Multiple Linear Regression
Multiplicative model
\[growth = intercept + temp + nitro + temp\times nitro\]
\[y_i=\beta_0+\beta_1x_{i1}+\beta_2x_{i2}+\beta_3x_{i1}x_{i2}+...+\epsilon_i\]
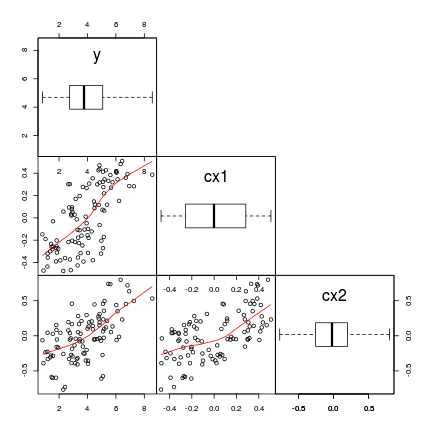
Assumtions
- normality, homogeneity of variance, linearity
- (multi)collinearity
Multiple Linear Regression
Variance inflation
\[ var.inf = \frac{1}{1-R^2} \] Collinear when \(var.inf >= 5\)
Some prefer \(>3\)
Worked Examples
Worked examples
| Format of loyn.csv data file | |||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|

|
loyn <- read.csv('../data/loyn.csv', strip.white=T)
head(loyn) ABUND AREA YR.ISOL DIST LDIST GRAZE ALT
1 5.3 0.1 1968 39 39 2 160
2 2.0 0.5 1920 234 234 5 60
3 1.5 0.5 1900 104 311 5 140
4 17.1 1.0 1966 66 66 3 160
5 13.8 1.0 1918 246 246 5 140
6 14.1 1.0 1965 234 285 3 130Worked Examples
Question: what effects do fragmentation variables have on the abundance of forest birds
Linear model:
\[
\begin{align}
Abund_i &\sim{} \mathcal{N}(\mu, \sigma^2)\\
\mu &= \beta_0 + \sum^N_{j=1:n}{\beta_j X_{ji}}\\
\beta_0, \beta_j &\sim{} \mathcal{N}(0, 1000)\\
\sigma &\sim{} Cauchy(0,5)\\
\end{align}
\]
Worked Examples
| Format of paruelo.csv data file | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|

|
paruelo <- read.csv('../data/paruelo.csv', strip.white=T)
head(paruelo) C3 LAT LONG MAP MAT JJAMAP DJFMAP
1 0.65 46.40 119.55 199 12.4 0.12 0.45
2 0.65 47.32 114.27 469 7.5 0.24 0.29
3 0.76 45.78 110.78 536 7.2 0.24 0.20
4 0.75 43.95 101.87 476 8.2 0.35 0.15
5 0.33 46.90 102.82 484 4.8 0.40 0.14
6 0.03 38.87 99.38 623 12.0 0.40 0.11Worked Examples
Question: what effects do fragmentation geographical variables have on the abundance of C3 grasses
Linear model:
\[
\begin{align}
\sqrt{C3_i} &\sim{} \mathcal{N}(\mu, \sigma^2)\\
\mu &= \beta_0 + \sum^N_{j=1:n}{\beta_j X_{ji}}\\
\beta_0, \beta_j &\sim{} \mathcal{N}(0, 1000)\\
\sigma &\sim{} Cauchy(0,5)\\
\end{align}
\]