Linear modelling assumptions
\[ \begin{align*} y_{i} &= \beta_0 + \beta_1 \times x_{i} + \varepsilon_i\\ \epsilon_i&\sim\mathcal{N}(0, \sigma^2) \\ \end{align*} \]
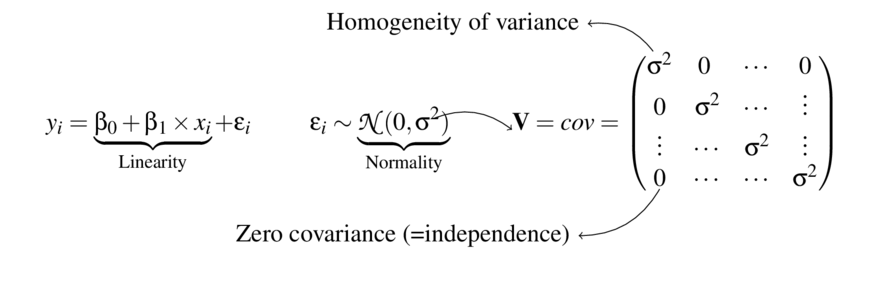
\[ \begin{align*} y_{i} &= \beta_0 + \beta_1 \times x_{i} + \varepsilon_i\\ \epsilon_i&\sim\mathcal{N}(0, \sigma^2) \\ \end{align*} \]

\[ \mathbf{V} = \underbrace{ \begin{pmatrix} \sigma^2&0&\cdots&0\\ 0&\sigma^2&\cdots&\vdots\\ \vdots&\cdots&\sigma^2&\vdots\\ 0&\cdots&\cdots&\sigma^2\\ \end{pmatrix}}_\text{Variance-covariance matrix} \]
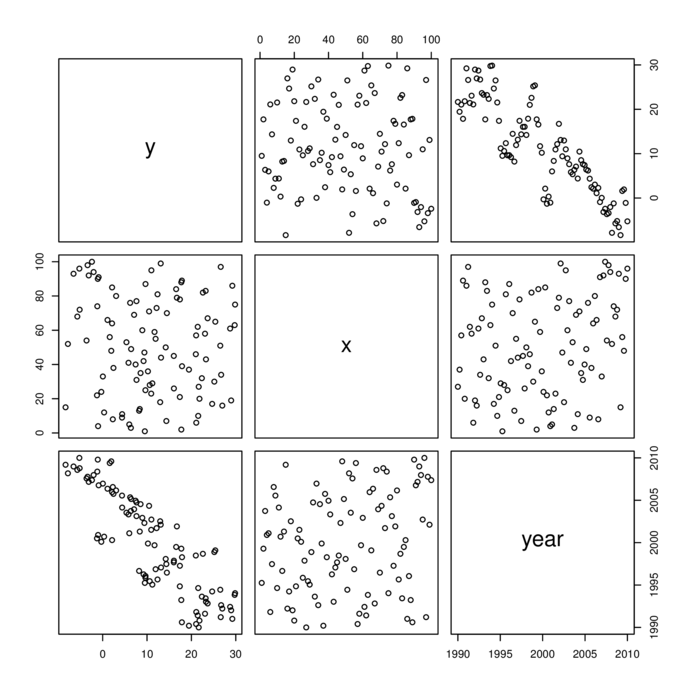
> data.t.lm <- lm(y~x, data=data.t)
> par(mfrow=c(2,3))
> plot(data.t.lm, which=1:6, ask=FALSE)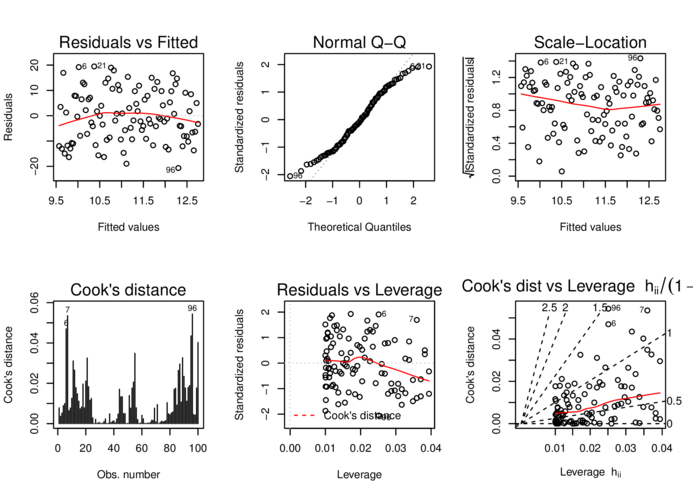
> acf(rstandard(data.t.lm))
> plot(rstandard(data.t.lm)~data.t$year)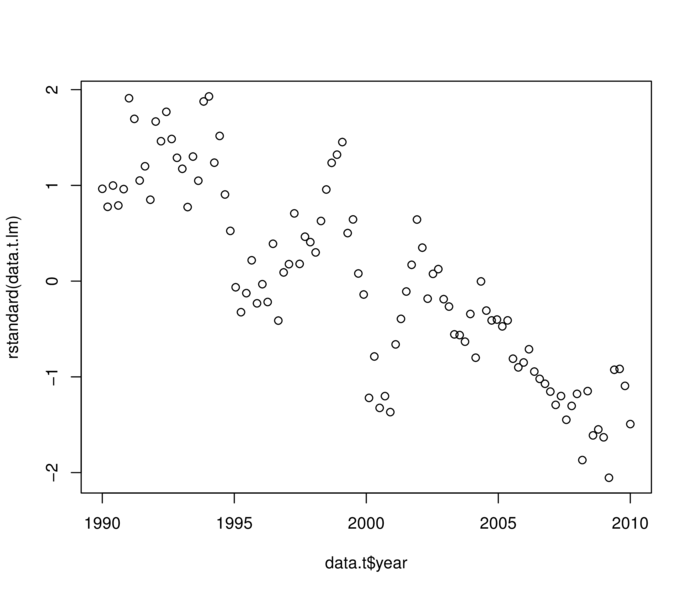
> library(car)
> vif(lm(y~x+year, data=data.t)) x year
1.040037 1.040037 > data.t.lm1 <- lm(y~x+year, data.t)> par(mfrow=c(1,2))
> plot(rstandard(data.t.lm1)~fitted(data.t.lm1))
> plot(rstandard(data.t.lm1)~data.t$year)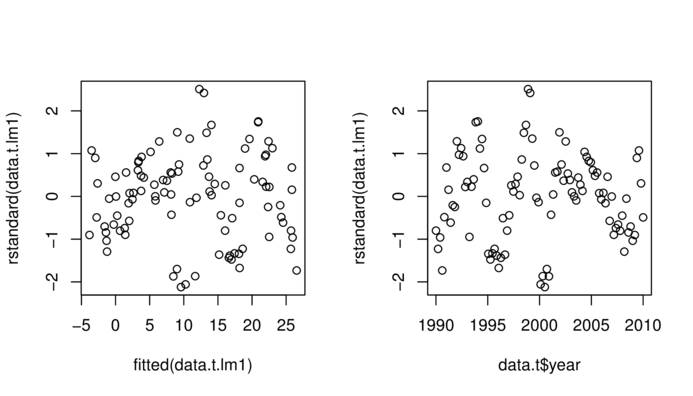
> acf(rstandard(data.t.lm1), lag=40)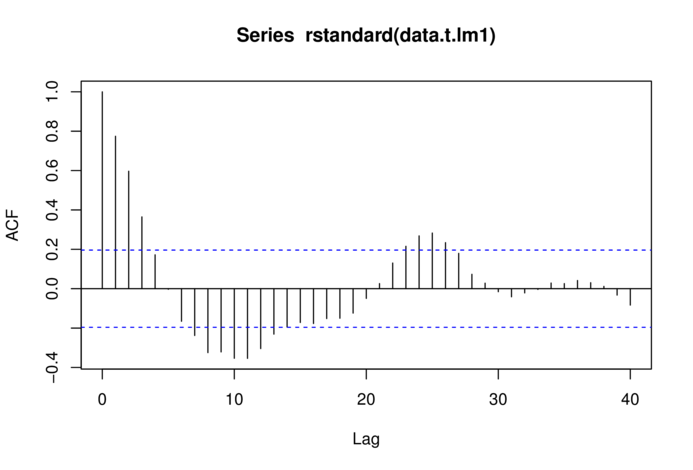
where:
> library(nlme)
> data.t.gls <- gls(y~x+year, data=data.t, method='REML')
> data.t.gls1 <- gls(y~x+year, data=data.t,
+ correlation=corAR1(form=~year),method='REML')> par(mfrow=c(1,2))
> plot(residuals(data.t.gls, type="normalized")~
+ fitted(data.t.gls))
> plot(residuals(data.t.gls1, type="normalized")~
+ fitted(data.t.gls1))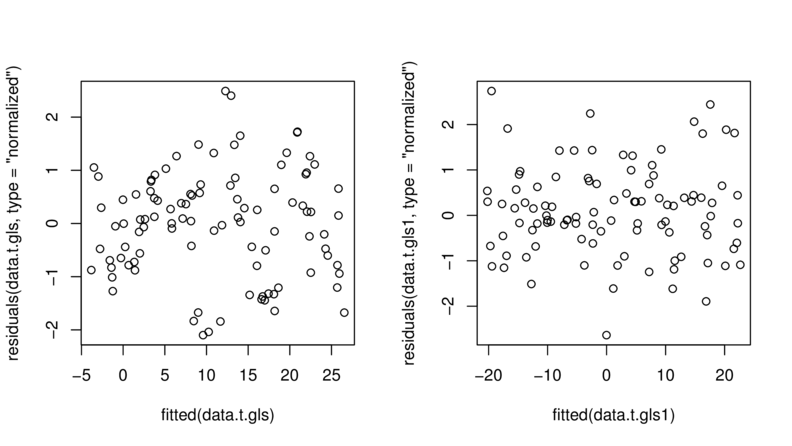
> par(mfrow=c(1,2))
> plot(residuals(data.t.gls, type="normalized")~
+ data.t$x)
> plot(residuals(data.t.gls1, type="normalized")~
+ data.t$x)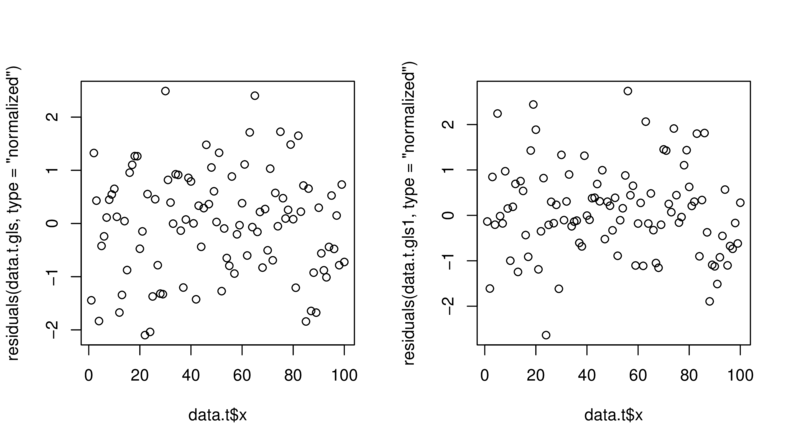
> par(mfrow=c(1,2))
> plot(residuals(data.t.gls, type="normalized")~
+ data.t$year)
> plot(residuals(data.t.gls1, type="normalized")~
+ data.t$year)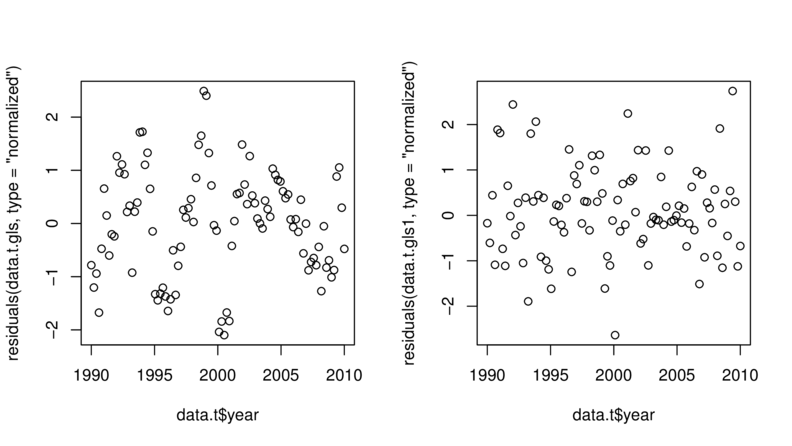
> par(mfrow=c(1,2))
> acf(residuals(data.t.gls, type='normalized'), lag=40)
> acf(residuals(data.t.gls1, type='normalized'), lag=40)
> AIC(data.t.gls, data.t.gls1) df AIC
data.t.gls 4 626.3283
data.t.gls1 5 536.7467> anova(data.t.gls, data.t.gls1) Model df AIC BIC logLik Test L.Ratio p-value
data.t.gls 1 4 626.3283 636.6271 -309.1642
data.t.gls1 2 5 536.7467 549.6203 -263.3734 1 vs 2 91.58158 <.0001> data.t.gls2 <- update(data.t.gls,
+ correlation=corARMA(form=~year,p=2,q=0))
> data.t.gls3 <- update(data.t.gls,
+ correlation=corARMA(form=~year,p=3,q=0))
> AIC(data.t.gls, data.t.gls1, data.t.gls2, data.t.gls3) df AIC
data.t.gls 4 626.3283
data.t.gls1 5 536.7467
data.t.gls2 6 538.1032
data.t.gls3 7 538.8376> summary(data.t.gls1)Generalized least squares fit by REML
Model: y ~ x + year
Data: data.t
AIC BIC logLik
536.7467 549.6203 -263.3734
Correlation Structure: ARMA(1,0)
Formula: ~year
Parameter estimate(s):
Phi1
0.9126603
Coefficients:
Value Std.Error t-value p-value
(Intercept) 4388.568 1232.6129 3.560378 0.0006
x 0.028 0.0086 3.296648 0.0014
year -2.195 0.6189 -3.545955 0.0006
Correlation:
(Intr) x
x 0.009
year -1.000 -0.010
Standardized residuals:
Min Q1 Med Q3 Max
-1.423389 1.710551 3.377925 4.372772 6.624381
Residual standard error: 3.37516
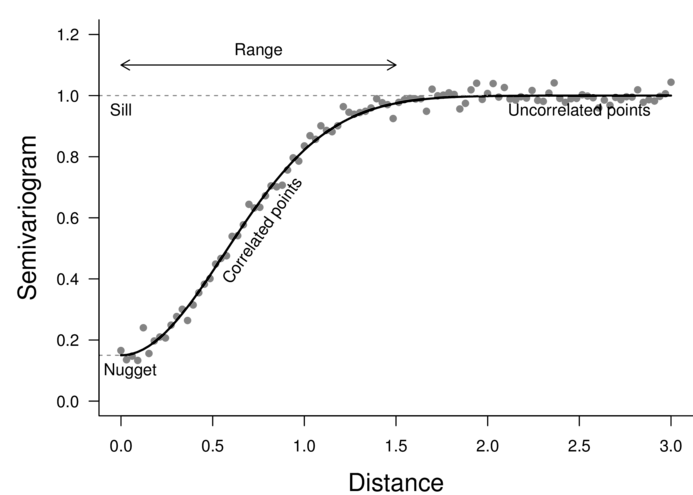
Degrees of freedom: 100 total; 97 residual2d Euclidean dissimilarity
Exponential decay
> library(sp)
> coordinates(data.s) <- ~LAT+LONG
> bubble(data.s,'y')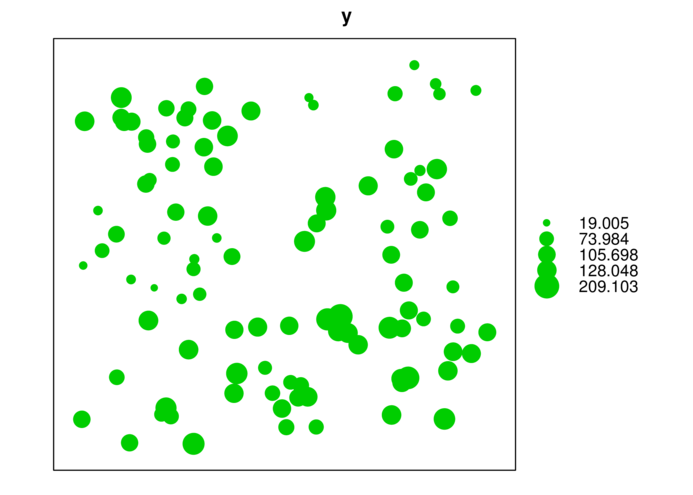
> data.s.lm <- lm(y~x, data=data.s)
> par(mfrow=c(2,3))
> plot(data.s.lm, which=1:6, ask=FALSE)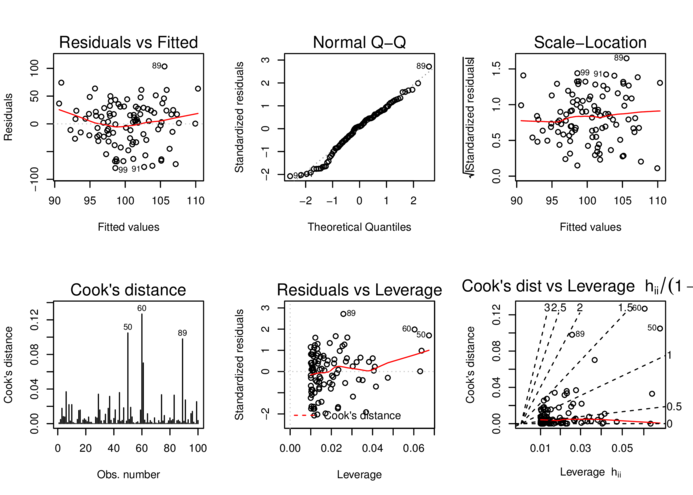
> data.s$Resid <- rstandard(data.s.lm)
> library(sp)
> #coordinates(data.s) <- ~LAT+LONG
> bubble(data.s,'Resid')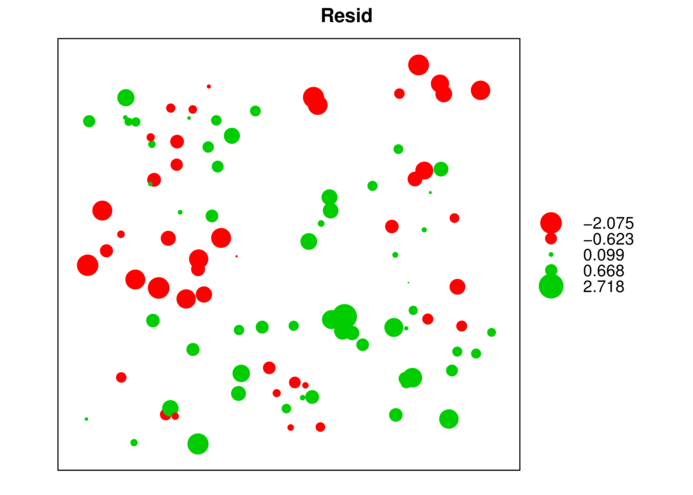
binnedaccording to distance and orientation (N)
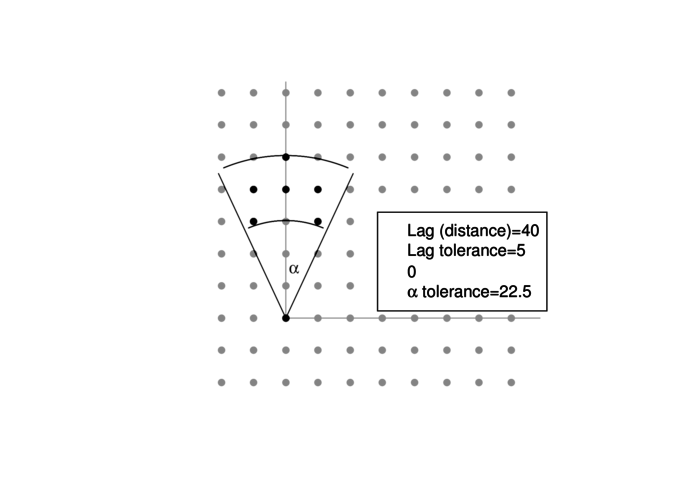
> library(nlme)
> data.s.gls <- gls(y~x, data.s, method='REML')
> plot(nlme:::Variogram(data.s.gls, form=~LAT+LONG,
+ resType="normalized"))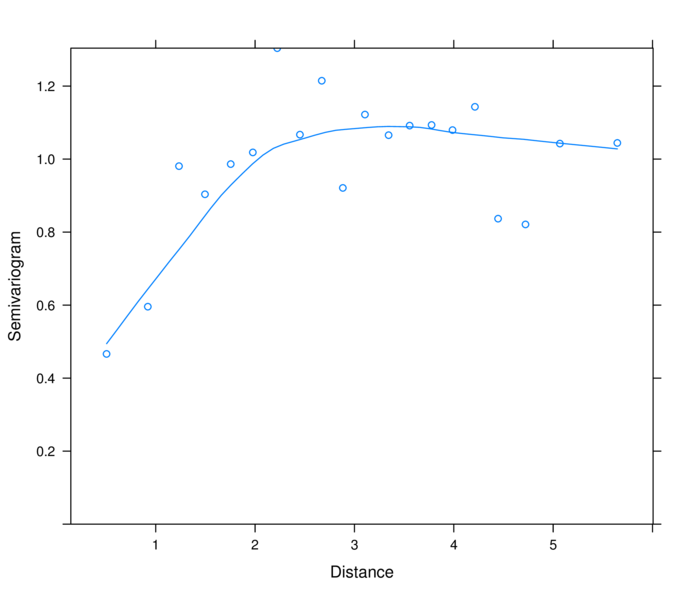
> library(gstat)
> plot(variogram(residuals(data.s.gls,"normalized")~1,
+ data=data.s, cutoff=6))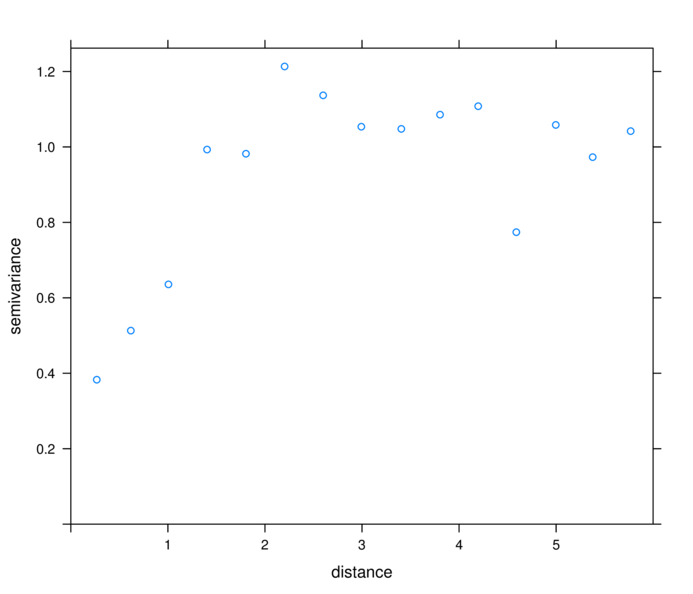
> library(gstat)
> plot(variogram(residuals(data.s.gls,"normalized")~1,
+ data=data.s, cutoff=6,alpha=c(0,45,90,135)))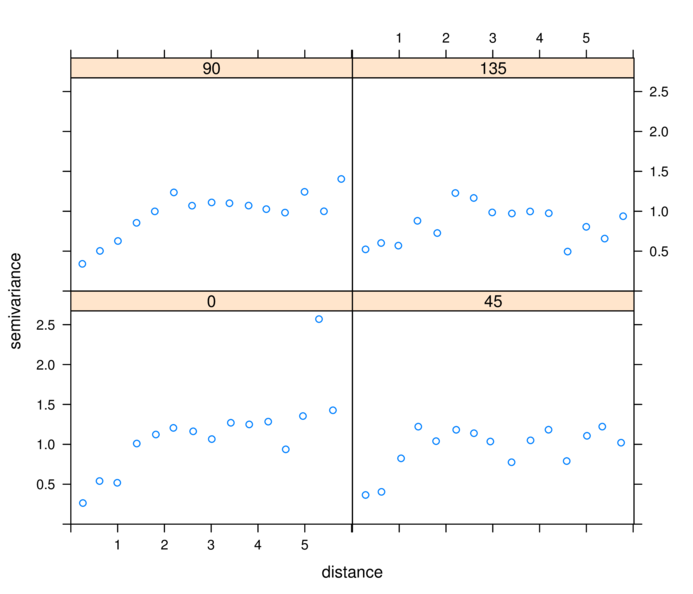
| correlation function | Correlation structure | Description |
|---|---|---|
| corExp(form=~lat+long) | Exponential | \(\Phi = 1-e^{-D/\rho}\) |
| corGaus(form=~lat+long) | Gaussian | \(\Phi = 1-e^{-(D/\rho)^2}\) |
| corLin(form=~lat+long) | Linear | \(\Phi = 1-(1-D/\rho)I(d<\rho)\) |
| corRatio(form=~lat+long) | Rational quadratic | \(\Phi = (d/\rho)^2/(1+(d/\rho)2)\) |
| corSpher(form=~lat+long) | Spherical | \(\Phi = 1-(1-1.5(d/\rho) + 0.5(d/\rho)^3)I(d<\rho)\) |
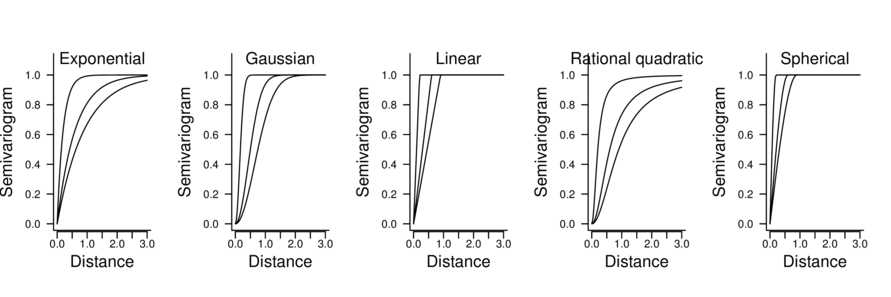
> data.s.glsExp <- update(data.s.gls,
+ correlation=corExp(form=~LAT+LONG, nugget=TRUE))
> data.s.glsGaus <- update(data.s.gls,
+ correlation=corGaus(form=~LAT+LONG, nugget=TRUE))
> #data.s.glsLin <- update(data.s/gls,
> # correlation=corLin(form=~LAT+LONG, nugget=TRUE))
> data.s.glsRatio <- update(data.s.gls,
+ correlation=corRatio(form=~LAT+LONG, nugget=TRUE))
> data.s.glsSpher <- update(data.s.gls,
+ correlation=corSpher(form=~LAT+LONG, nugget=TRUE))
>
> AIC(data.s.gls, data.s.glsExp, data.s.glsGaus, data.s.glsRatio, data.s.glsSpher) df AIC
data.s.gls 3 1013.9439
data.s.glsExp 5 974.3235
data.s.glsGaus 5 976.4422
data.s.glsRatio 5 974.7862
data.s.glsSpher 5 975.5244> plot(residuals(data.s.glsExp, type="normalized")~
+ fitted(data.s.glsExp))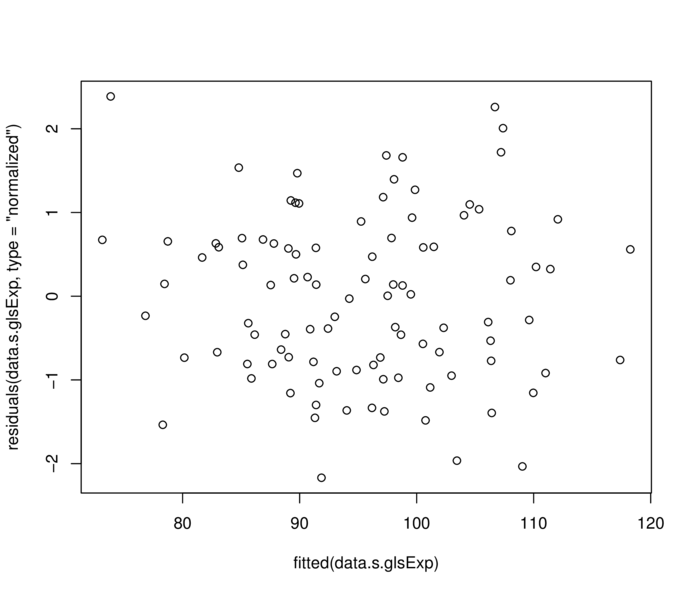
> plot(nlme:::Variogram(data.s.glsExp, form=~LAT+LONG,
+ resType="normalized"))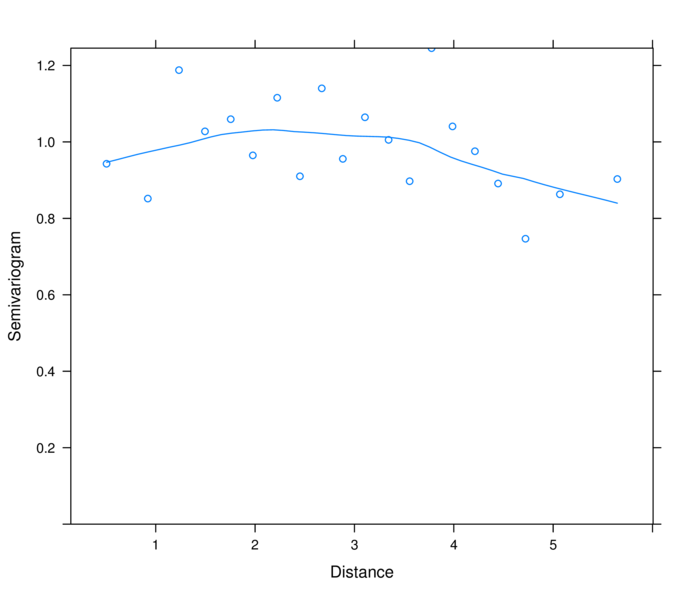
> summary(data.s.glsExp)Generalized least squares fit by REML
Model: y ~ x
Data: data.s
AIC BIC logLik
974.3235 987.2484 -482.1618
Correlation Structure: Exponential spatial correlation
Formula: ~LAT + LONG
Parameter estimate(s):
range nugget
1.6956723 0.1280655
Coefficients:
Value Std.Error t-value p-value
(Intercept) 65.90018 21.824752 3.019516 0.0032
x 0.94572 0.286245 3.303886 0.0013
Correlation:
(Intr)
x -0.418
Standardized residuals:
Min Q1 Med Q3 Max
-1.6019483 -0.3507695 0.1608776 0.6451751 2.1331505
Residual standard error: 47.68716
Degrees of freedom: 100 total; 98 residual> anova(data.s.glsExp)Denom. DF: 98
numDF F-value p-value
(Intercept) 1 23.45923 <.0001
x 1 10.91566 0.0013> xs <- seq(min(data.s$x), max(data.s$x), l=100)
> xmat <- model.matrix(~x, data.frame(x=xs))
>
> mpred <- function(model, xmat, data.s) {
+ pred <- as.vector(coef(model) %*% t(xmat))
+ (se<-sqrt(diag(xmat %*% vcov(model) %*% t(xmat))))
+ ci <- data.frame(pred+outer(se,qt(df=nrow(data.s)-2,c(.025,.975))))
+ colnames(ci) <- c('lwr','upr')
+ data.s.sum<-data.frame(pred, x=xs,se,ci)
+ data.s.sum
+ }
>
> data.s.sum<-mpred(data.s.glsExp, xmat, data.s)
> data.s.sum$resid <- data.s.sum$pred+residuals(data.s.glsExp)
>
> library(ggplot2)
> ggplot(data.s.sum, aes(y=pred, x=x))+
+ geom_ribbon(aes(ymin=lwr, ymax=upr), fill='blue', alpha=0.2) +
+ geom_line()+geom_point(aes(y=resid)) + theme_classic()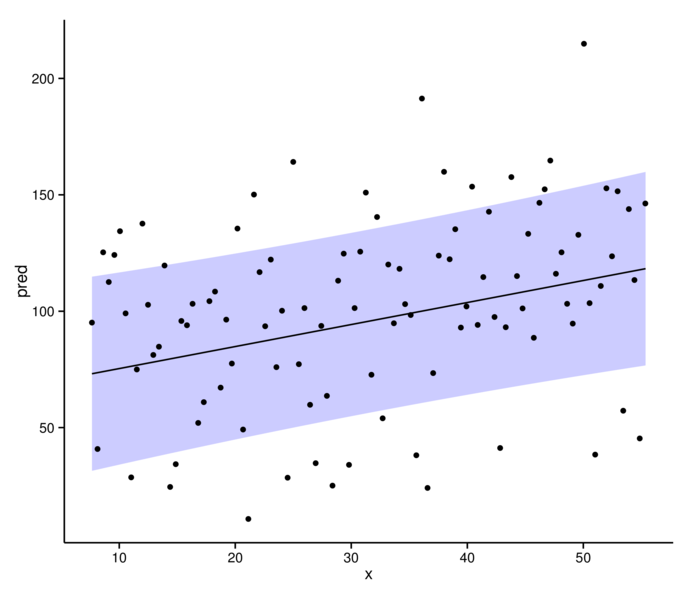
> #plot(pred~x, ylim=c(min(lwr), max(upr)), data.s.sum, type="l")
> #lines(lwr~x, data.s.sum)
> #lines(upr~x, data.s.sum)
> #points(pred+residuals(data.s.glsExp)~x, data.s.sum)
>
> #newdata <- data.frame(x=xs)[1,]