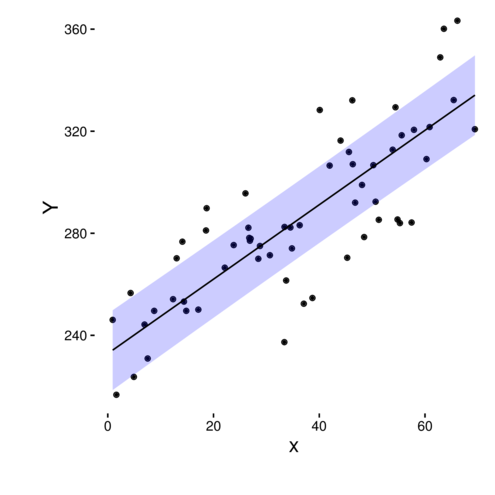
Linear models
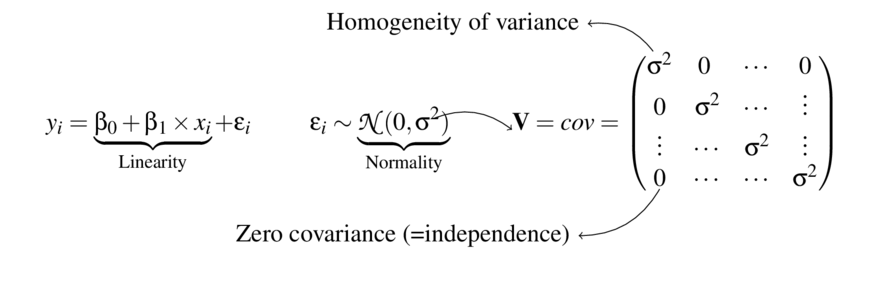
How maximize power?

How maximize power?

How maximize power?
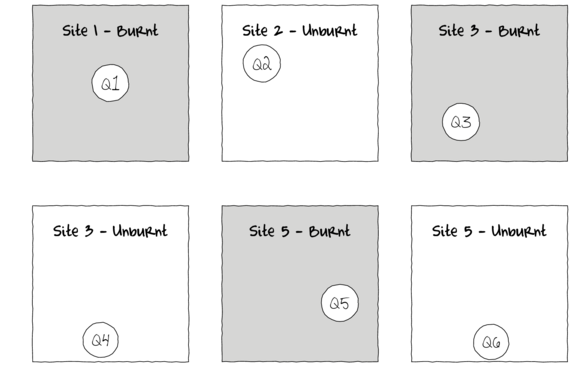
To increase power - without more sites (replicates)
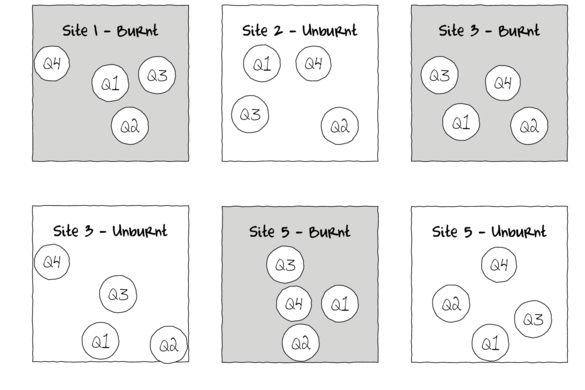
Subreplicates - yet not independent



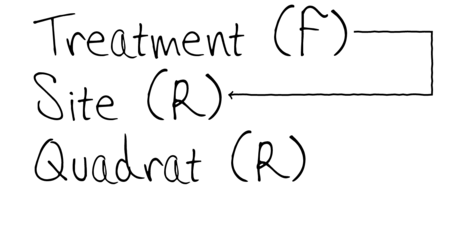
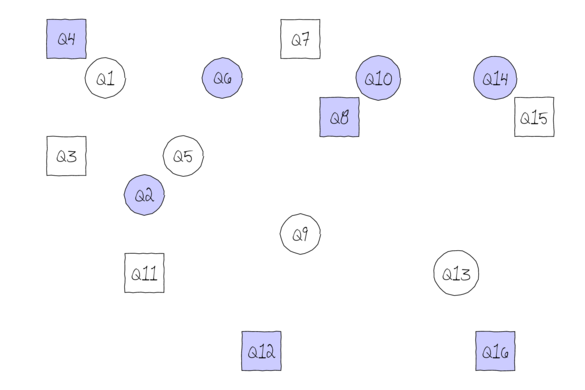
To increase power…
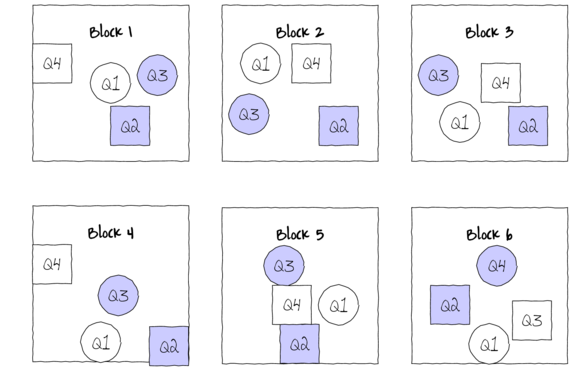
Block treatments together - yet not independent



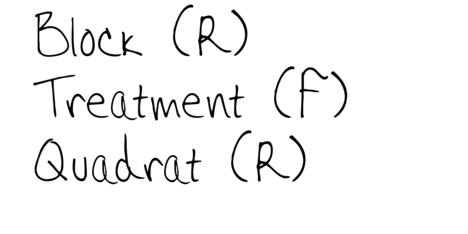

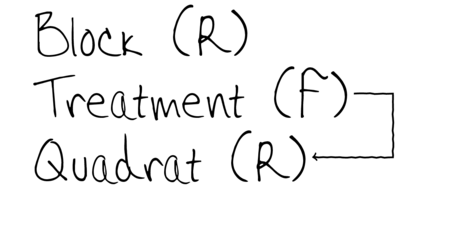


> data.rcb <- read.csv('../data/data.rcb.csv')
> head(data.rcb) y x block
1 281.1091 18.58561 Block1
2 295.6535 26.04867 Block1
3 328.3234 40.09974 Block1
4 360.1672 63.57455 Block1
5 276.7050 14.11774 Block1
6 348.9709 62.88728 Block1> library(ggplot2)
> ggplot(data.rcb, aes(y=y, x=x)) + geom_point() + geom_smooth(method='lm')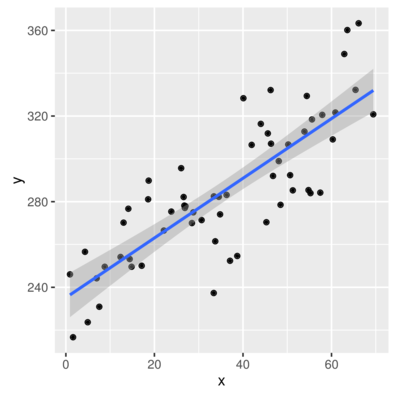
> library(ggplot2)
> ggplot(data.rcb, aes(y=y, x=x,color=block))+geom_point()+
+ geom_smooth(method='lm')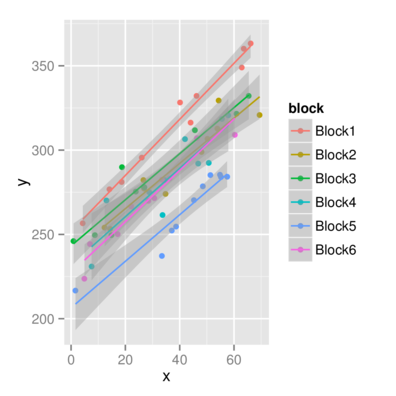
Simple linear regression - wrong
> data.rcb.lm <- lm(y~x, data.rcb)
Generalized least squares - more correct
> library(nlme)
> data.rcb.gls <- gls(y~x, data.rcb, method='REML')Model validation
> plot(data.rcb.gls)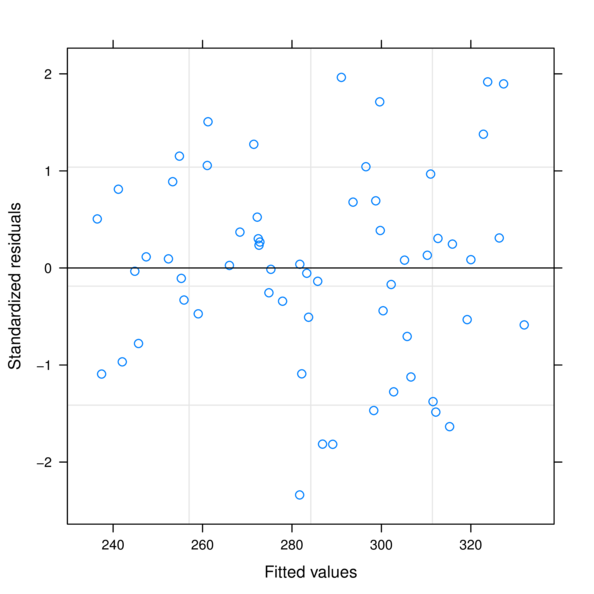
> plot(residuals(data.rcb.gls, type='normalized') ~
+ data.rcb$block)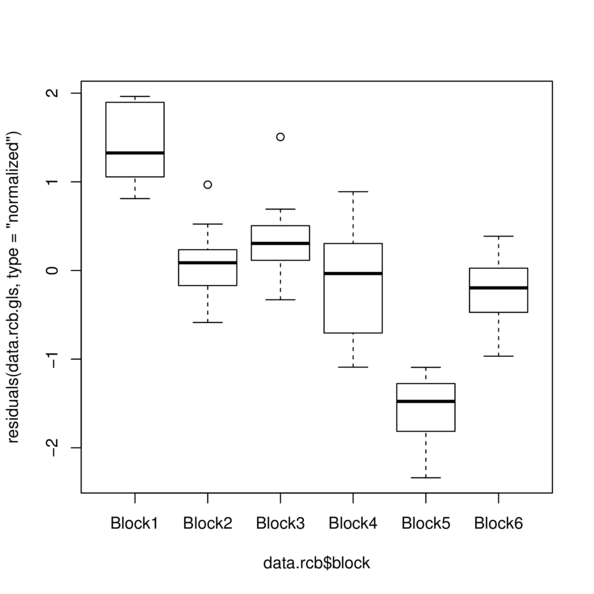
- So what about ANCOVA
> library(ggplot2)
> ggplot(data.rcb, aes(y=y, x=x, color=block))+
+ geom_smooth(method="lm")+geom_point()+theme_classic()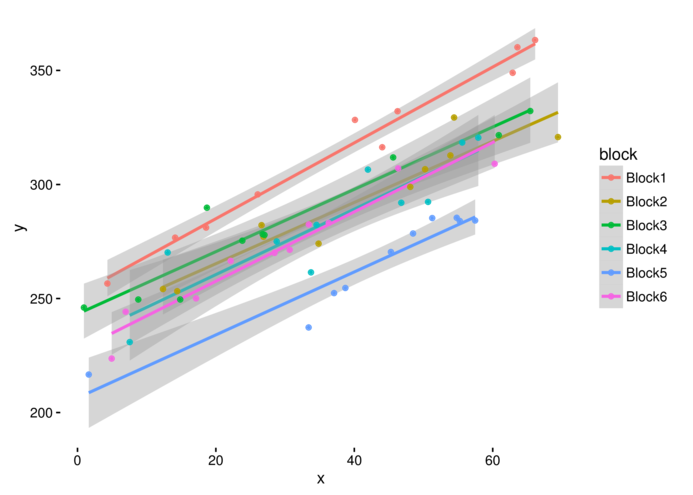
What if we add block as a predictor? (like ANCOVA)
> library(nlme)
> data.rcb.gls1 <- gls(y~x+block, data.rcb, method='REML')
> plot(data.rcb.gls)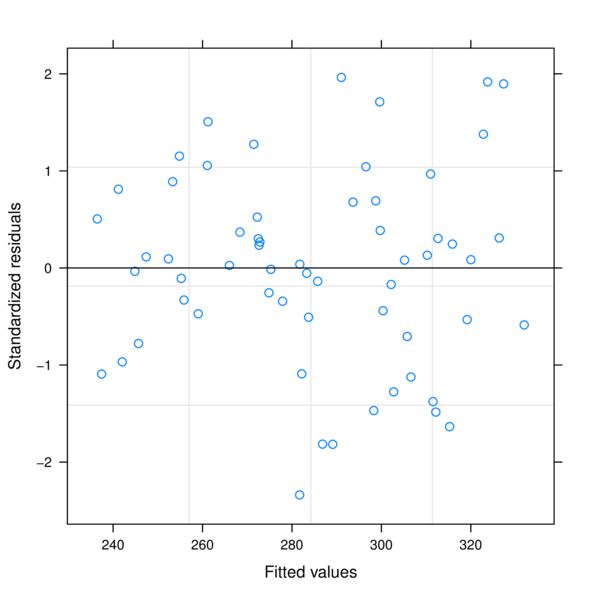
> plot(residuals(data.rcb.gls1, type='normalized') ~
+ data.rcb$block)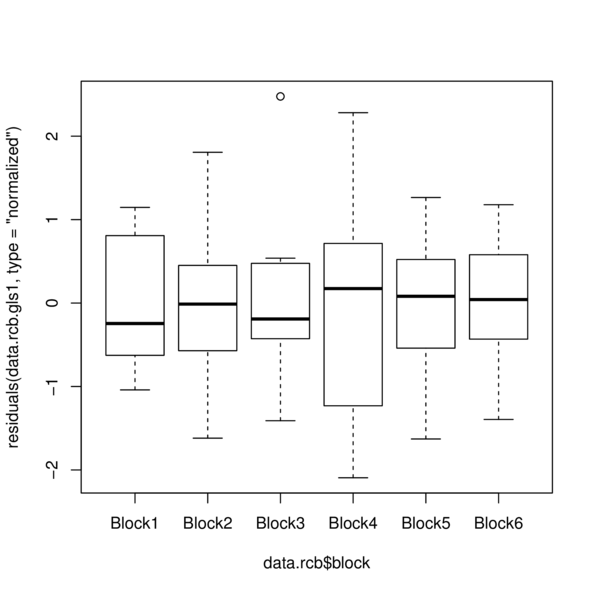
Model in dependency structure
> library(nlme)
> data.rcb.gls2<-gls(y~x,data.rcb,
+ correlation=corCompSymm(form=~1|block),
+ method="REML")
> plot(residuals(data.rcb.gls2, type='normalized') ~
+ fitted(data.rcb.gls2))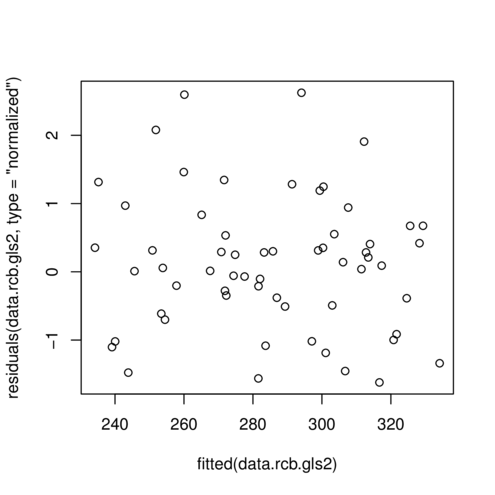
> plot(residuals(data.rcb.gls2, type='normalized') ~
+ data.rcb$block)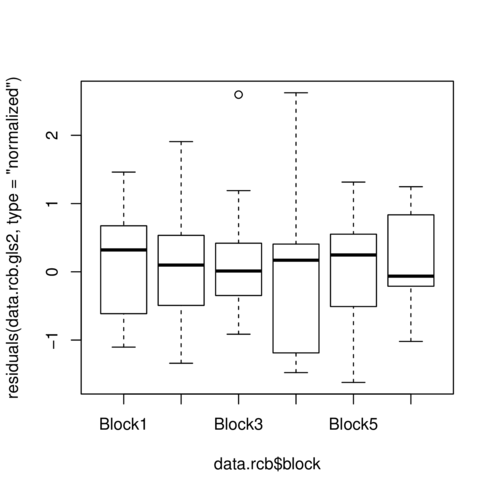
> summary(data.rcb.gls2)Generalized least squares fit by REML
Model: y ~ x
Data: data.rcb
AIC BIC logLik
458.9521 467.1938 -225.476
Correlation Structure: Compound symmetry
Formula: ~1 | block
Parameter estimate(s):
Rho
0.8052553
Coefficients:
Value Std.Error t-value p-value
(Intercept) 232.8193 7.823394 29.75937 0
x 1.4591 0.063789 22.87392 0
Correlation:
(Intr)
x -0.292
Standardized residuals:
Min Q1 Med Q3 Max
-2.19174920 -0.59481155 0.05261311 0.59571239 1.83321624
Residual standard error: 20.18017
Degrees of freedom: 60 total; 58 residual> data.rcb.lme <- lme(y~x, random=~1|block, data.rcb,
+ method='REML')
> plot(data.rcb.lme)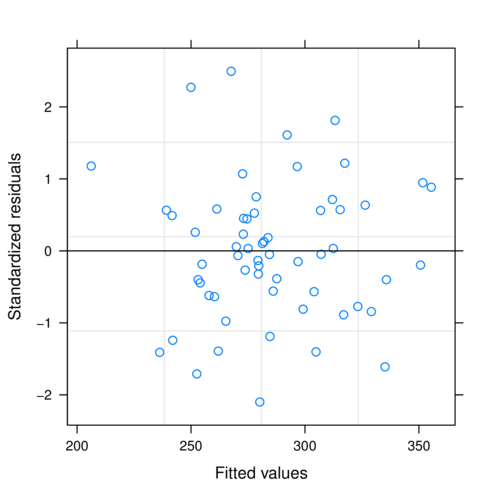
> plot(residuals(data.rcb.lme, type='normalized') ~ fitted(data.rcb.lme))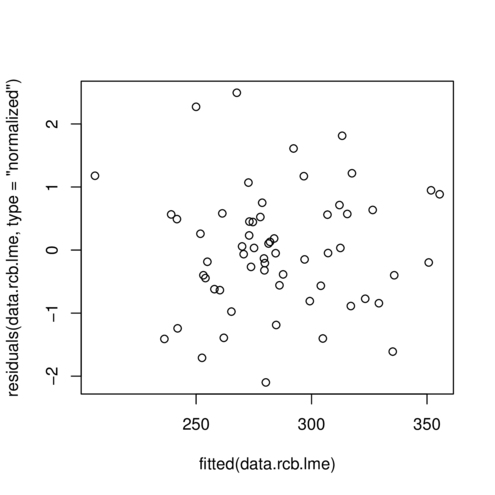
> plot(residuals(data.rcb.lme, type='normalized') ~ data.rcb$block)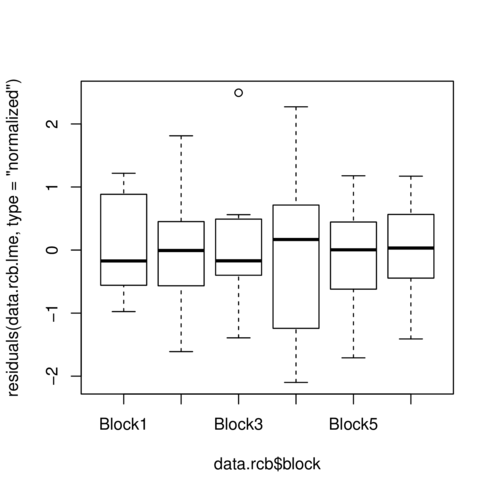
> summary(data.rcb.lme)Linear mixed-effects model fit by REML
Data: data.rcb
AIC BIC logLik
458.9521 467.1938 -225.476
Random effects:
Formula: ~1 | block
(Intercept) Residual
StdDev: 18.10888 8.905485
Fixed effects: y ~ x
Value Std.Error DF t-value p-value
(Intercept) 232.8193 7.823393 53 29.75937 0
x 1.4591 0.063789 53 22.87392 0
Correlation:
(Intr)
x -0.292
Standardized Within-Group Residuals:
Min Q1 Med Q3 Max
-2.09947262 -0.57994305 -0.04874031 0.56685096 2.49464217
Number of Observations: 60
Number of Groups: 6 > anova(data.rcb.lme) numDF denDF F-value p-value
(Intercept) 1 53 1452.2883 <.0001
x 1 53 523.2164 <.0001> intervals(data.rcb.lme)Approximate 95% confidence intervals
Fixed effects:
lower est. upper
(Intercept) 217.127551 232.819291 248.511031
x 1.331156 1.459101 1.587045
attr(,"label")
[1] "Fixed effects:"
Random Effects:
Level: block
lower est. upper
sd((Intercept)) 9.597555 18.10888 34.16822
Within-group standard error:
lower est. upper
7.361789 8.905485 10.772878 > vc<-as.numeric(as.matrix(VarCorr(data.rcb.lme))[,1])
> vc/sum(vc)[1] 0.8052553 0.1947447> library(effects)
> plot(allEffects(data.rcb.lme, partial.residuals=TRUE))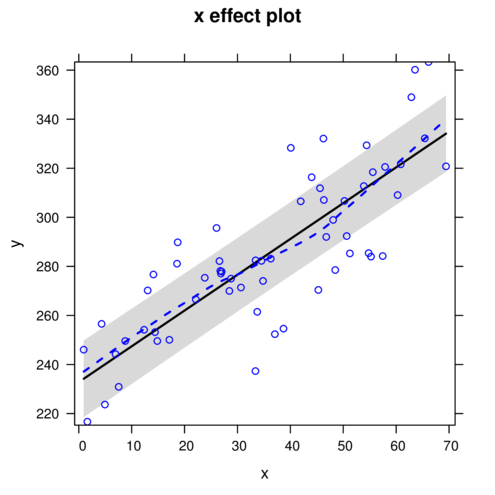
> predict(data.rcb.lme, newdata=data.frame(x=30:40),level=0) [1] 276.5923 278.0514 279.5105 280.9696 282.4287 283.8878 285.3469 286.8060 288.2651 289.7242
[11] 291.1833
attr(,"label")
[1] "Predicted values"> predict(data.rcb.lme, newdata=data.frame(x=30:40,
+ block='Block1'),level=1) Block1 Block1 Block1 Block1 Block1 Block1 Block1 Block1 Block1 Block1 Block1
302.7422 304.2013 305.6604 307.1195 308.5786 310.0377 311.4968 312.9559 314.4150 315.8741 317.3332
attr(,"label")
[1] "Predicted values"Step 1. gather model coefficients
> coefs <- fixef(data.rcb.lme)
> coefs(Intercept) x
232.819291 1.459101 Step 2. generate prediction model matrix
> xs <- seq(min(data.rcb$x), max(data.rcb$x), l=100)
> Xmat <- model.matrix(~x, data.frame(x=xs))
> head(Xmat) (Intercept) x
1 1 0.9373233
2 1 1.6292032
3 1 2.3210830
4 1 3.0129628
5 1 3.7048426
6 1 4.3967225Step 3. calculate predicted y
> ys <- t(coefs %*% t(Xmat))
> head(ys) [,1]
1 234.1869
2 235.1965
3 236.2060
4 237.2155
5 238.2250
6 239.2346Step 3. calculate confidence interval
> SE <- sqrt(diag(Xmat %*% vcov(data.rcb.lme) %*% t(Xmat)))
> CI <- 2*SE
> #OR
> CI <- qt(0.975,length(data.rcb$x)-2)*SE
> data.rcb.pred <- data.frame(x=xs, fit=ys, se=SE,
+ lower=ys-CI, upper=ys+CI)
> head(data.rcb.pred) x fit se lower upper
1 0.9373233 234.1869 7.806128 218.5613 249.8126
2 1.6292032 235.1965 7.793653 219.5958 250.7972
3 2.3210830 236.2060 7.781408 220.6298 251.7822
4 3.0129628 237.2155 7.769395 221.6634 252.7676
5 3.7048426 238.2250 7.757614 222.6965 253.7536
6 4.3967225 239.2346 7.746067 223.7291 254.7400Step 4. plot it
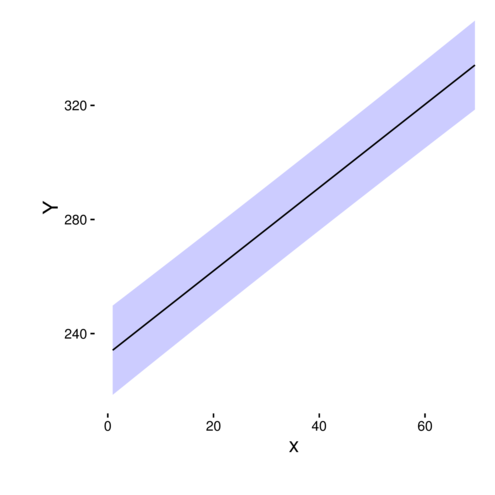
> library(ggplot2)
> ggplot(data.rcb.pred, aes(y=fit, x=x)) +
+ geom_ribbon(aes(ymin=lower,ymax=upper),fill='blue',alpha=0.2)+
+ geom_line()+
+ scale_y_continuous('Y')+
+ theme_classic()+
+ theme(axis.title.x=element_text(size=rel(1.25), vjust=-2),
+ axis.title.y=element_text(size=rel(1.25), vjust=2),
+ plot.margin=unit(c(0.1,0.1,2,2),'lines'))
>
> ## plot(fit~x, data=data.rcb.pred,type='n',axes=F, ann=F)
> ## points(y~x, data=data.rcb, pch=16, col='grey')
> ## with(data.rcb.pred, polygon(c(x,rev(x)), c(lower, rev(upper)),
> ## col="#0000FF50",border=FALSE))
> ## lines(fit~x,data=data.rcb.pred)
> ## lines(lower~x,data=data.rcb.pred, lty=2)
> ## lines(upper~x,data=data.rcb.pred, lty=2)
> ## axis(1)
> ## mtext('X',1,line=3)
> ## axis(2,las=1)
> ## mtext('Y',2,line=3)
> ## box(bty='l')Linear mixed effects model
Step 4. plot it (with partial observed values)
> data.rcb$res <- predict(data.rcb.lme, level=1)+
+ residuals(data.rcb.lme)
>
> library(ggplot2)
> ggplot(data.rcb.pred, aes(y=fit, x=x)) +
+ geom_point(data=data.rcb, aes(y=res))+
+ geom_ribbon(aes(ymin=lower,ymax=upper),fill='blue',alpha=0.2)+
+ geom_line()+
+ scale_y_continuous('Y')+
+ theme_classic()+
+ theme(axis.title.x=element_text(size=rel(1.25), vjust=-2),
+ axis.title.y=element_text(size=rel(1.25), vjust=2),
+ plot.margin=unit(c(0.1,0.1,2,2),'lines'))