RCB design
Simple
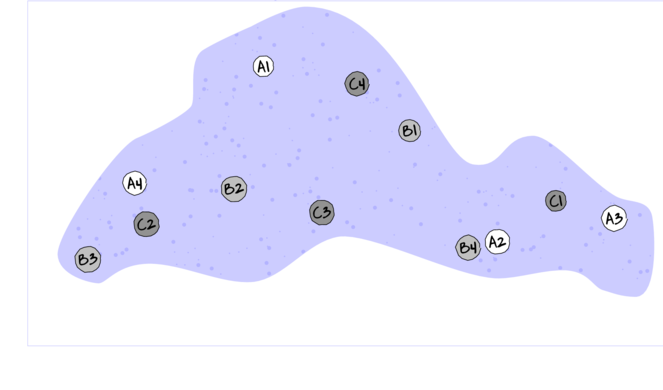
Randomized block design
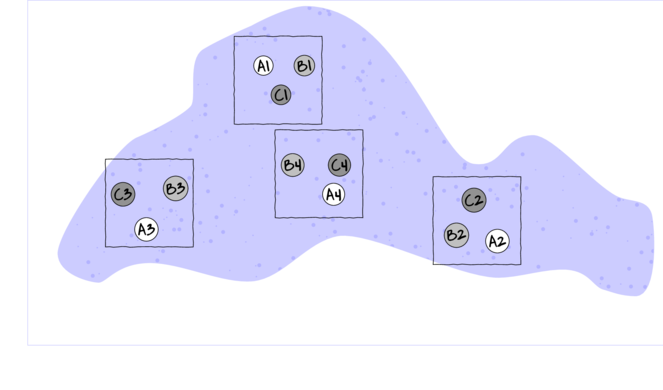
Simple

Randomized block design

\(y_{ij} = \mu + \beta_i + \alpha_j + \varepsilon_{ij}\)
\(\mu\) - the mean of the first treatment group
\(\beta\) - the random (Block) effect
\(\alpha\) - the main within Block effect
e.g.

| Single factor ANOVA | Variance-covariance | ||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
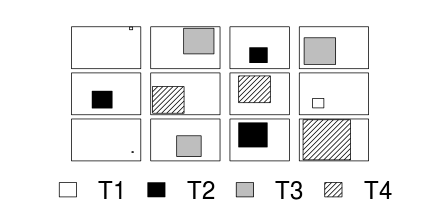
|
|
||||||||||||||||||||||||||
| Randomized Complete Block | Variance-covariance | ||||||||||||||||||||||||||
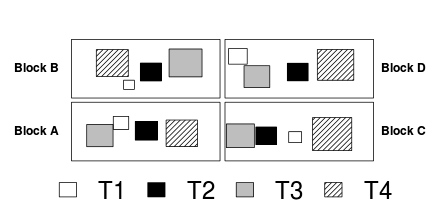
|
|
||||||||||||||||||||||||||
| Repeated Measures Design | Variance-covariance | ||||||||||||||||||||||||||
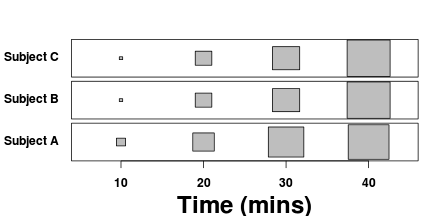
|
|
||||||||||||||||||||||||||
> data.rcb1 <- read.csv('../data/data.rcb1.csv', strip.white=TRUE)
> head(data.rcb1) y A Block
1 37.39761 A B1
2 61.47033 B B1
3 78.07370 C B1
4 30.59803 A B2
5 59.00035 B B2
6 76.72575 C B2Normality and homogeneity of variance
> boxplot(y~A, data.rcb1)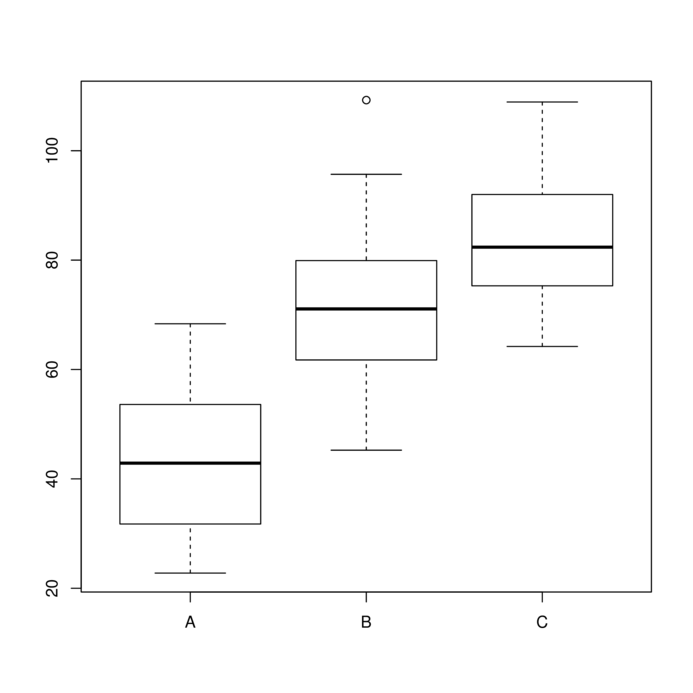
No block by within-block interaction
> library(ggplot2)
> ggplot(data.rcb1, aes(y=y, x=A, group=Block,color=Block)) +
+ geom_line() +
+ guides(color=guide_legend(ncol=3))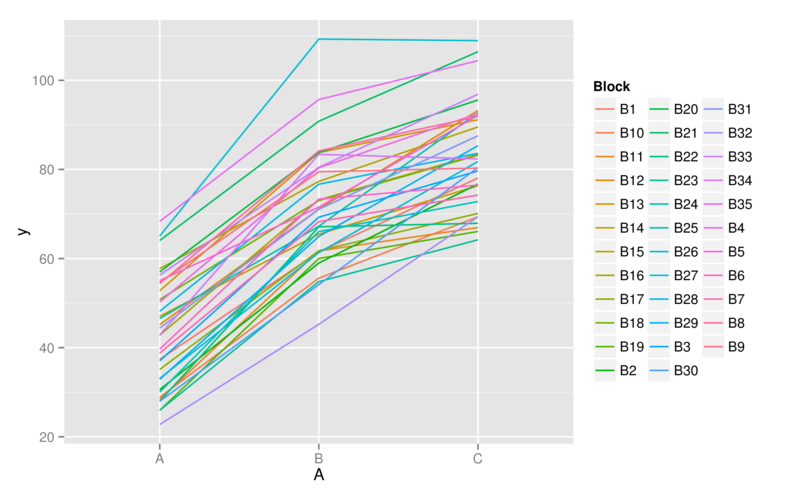
No block by within-block interaction
> library(car)
> residualPlots(lm(y~Block+A, data.rcb1))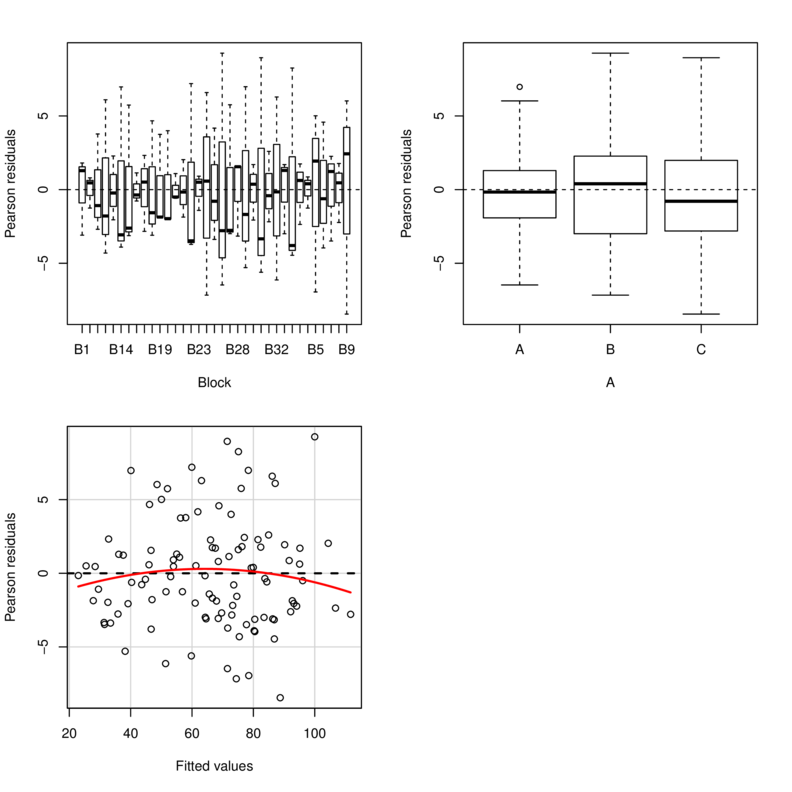
Test stat Pr(>|t|)
Block NA NA
A NA NA
Tukey test -0.885 0.376> library(nlme)
> data.rcb1.lme <- lme(y~A, random=~1|Block,
+ data=data.rcb1)
> acf(resid(data.rcb1.lme))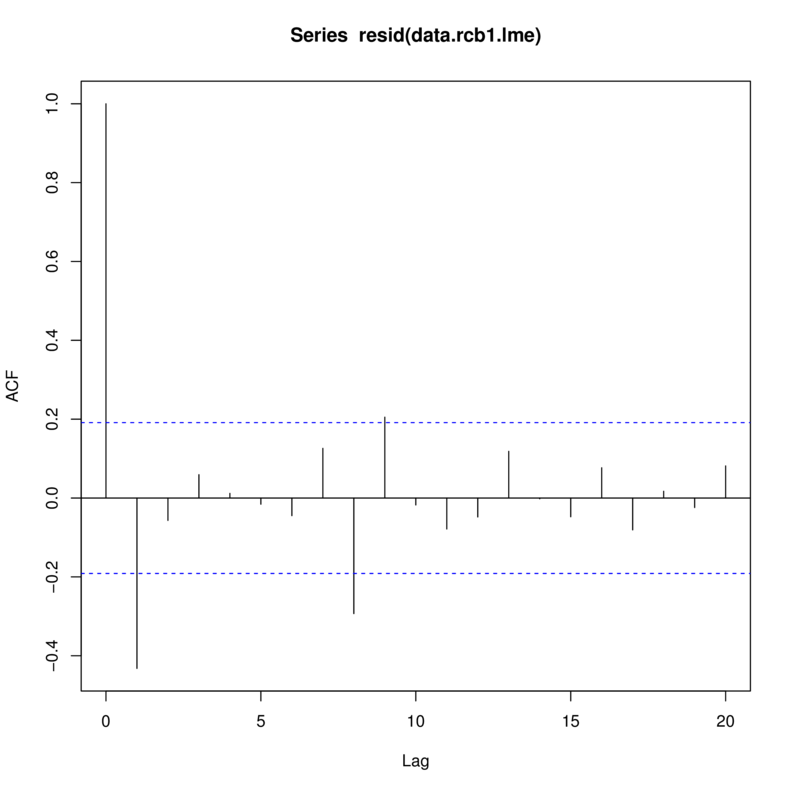
> #Assuming sphericity
> data.rcb1.lme <- lme(y~A, random=~1|Block, data=data.rcb1,
+ method='REML')
> data.rcb1.lme1 <- lme(y~A, random=~A|Block, data=data.rcb1,
+ method='REML')
> AIC(data.rcb1.lme, data.rcb1.lme1) df AIC
data.rcb1.lme 5 722.1087
data.rcb1.lme1 10 727.2001> anova(data.rcb1.lme, data.rcb1.lme1) Model df AIC BIC logLik Test L.Ratio p-value
data.rcb1.lme 1 5 722.1087 735.2336 -356.0544
data.rcb1.lme1 2 10 727.2001 753.4499 -353.6001 1 vs 2 4.908574 0.4271> #Assuming sphericity
> data.rcb1.lme.AR1 <- lme(y~A, random=~1|Block, data=data.rcb1,
+ correlation=corAR1(),method='REML')
> data.rcb1.lme1.AR1 <- lme(y~A, random=~A|Block, data=data.rcb1,
+ correlation=corAR1(),method='REML')
> AIC(data.rcb1.lme, data.rcb1.lme1,data.rcb1.lme.AR1, data.rcb1.lme1.AR1) df AIC
data.rcb1.lme 5 722.1087
data.rcb1.lme1 10 727.2001
data.rcb1.lme.AR1 6 723.3178
data.rcb1.lme1.AR1 11 729.2001> anova(data.rcb1.lme, data.rcb1.lme1,data.rcb1.lme.AR1, data.rcb1.lme1.AR1) Model df AIC BIC logLik Test L.Ratio p-value
data.rcb1.lme 1 5 722.1087 735.2336 -356.0544
data.rcb1.lme1 2 10 727.2001 753.4499 -353.6001 1 vs 2 4.908574 0.4271
data.rcb1.lme.AR1 3 6 723.3178 739.0676 -355.6589 2 vs 3 4.117606 0.3903
data.rcb1.lme1.AR1 4 11 729.2001 758.0748 -353.6001 3 vs 4 4.117606 0.5326> plot(data.rcb1.lme)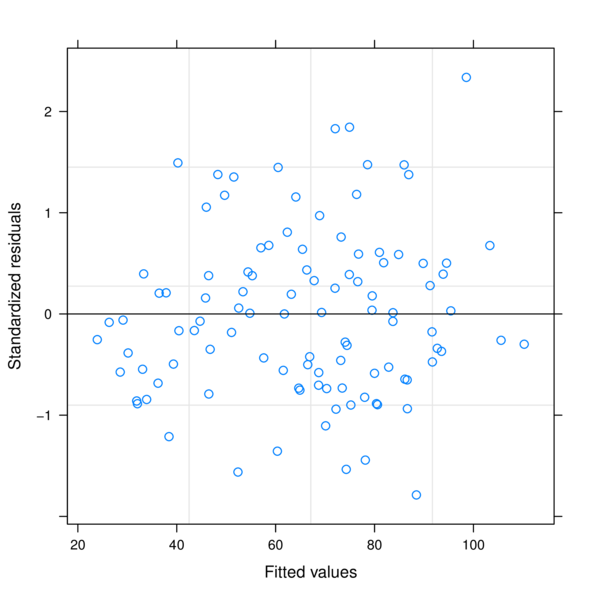
> plot(resid(data.rcb1.lme, type='normalized') ~
+ data.rcb1$A)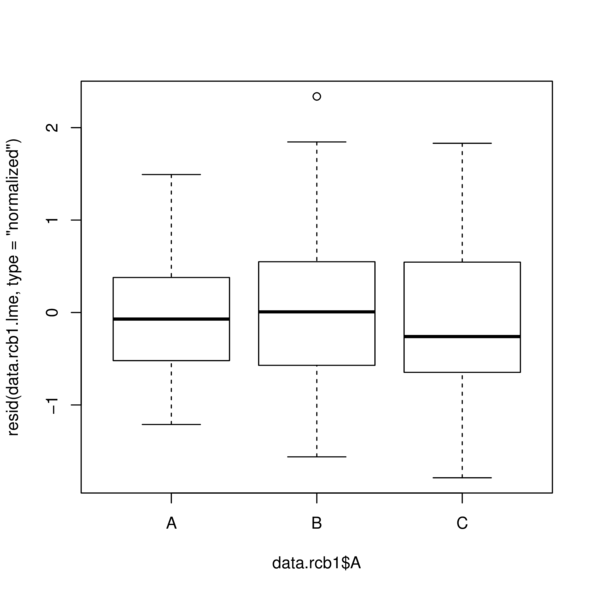
> library(effects)
> plot(Effect('A',data.rcb1.lme))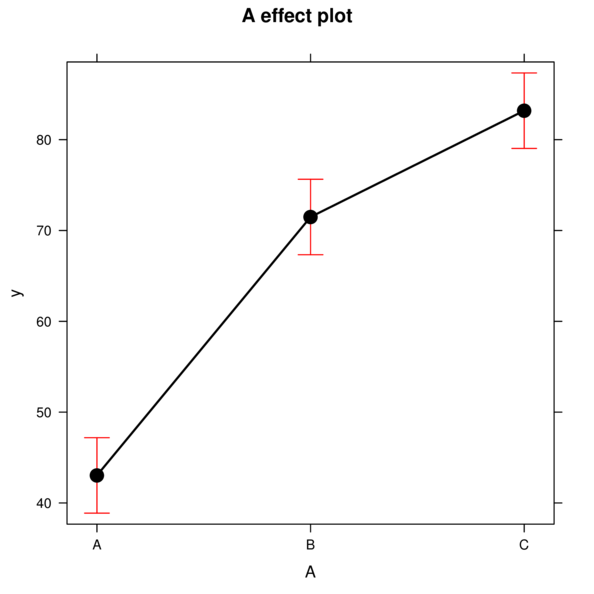
> summary(data.rcb1.lme)Linear mixed-effects model fit by REML
Data: data.rcb1
AIC BIC logLik
722.1087 735.2336 -356.0544
Random effects:
Formula: ~1 | Block
(Intercept) Residual
StdDev: 11.51409 4.572284
Fixed effects: y ~ A
Value Std.Error DF t-value p-value
(Intercept) 43.03434 2.094074 68 20.55053 0
AB 28.45241 1.092985 68 26.03185 0
AC 40.15556 1.092985 68 36.73936 0
Correlation:
(Intr) AB
AB -0.261
AC -0.261 0.500
Standardized Within-Group Residuals:
Min Q1 Med Q3 Max
-1.78748258 -0.57867597 -0.07108159 0.49990644 2.33727672
Number of Observations: 105
Number of Groups: 35 > intervals(data.rcb1.lme)Approximate 95% confidence intervals
Fixed effects:
lower est. upper
(Intercept) 38.85568 43.03434 47.21300
AB 26.27140 28.45241 30.63343
AC 37.97455 40.15556 42.33658
attr(,"label")
[1] "Fixed effects:"
Random Effects:
Level: Block
lower est. upper
sd((Intercept)) 8.964236 11.51409 14.78925
Within-group standard error:
lower est. upper
3.864944 4.572284 5.409077 > VarCorr(data.rcb1.lme)Block = pdLogChol(1)
Variance StdDev
(Intercept) 132.57434 11.514093
Residual 20.90578 4.572284> anova(data.rcb1.lme) numDF denDF F-value p-value
(Intercept) 1 68 1089.3799 <.0001
A 2 68 714.0295 <.0001[1] 0.6516126[1] 0.300933[1] 0.04745443[1] 0.9525456> library(lme4)
> data.rcb1.lmer <- lmer(y~A+(1|Block), data=data.rcb1, REML=TRUE,
+ control=lmerControl(check.nobs.vs.nRE="ignore"))
> data.rcb1.lmer1 <- lmer(y~A+(A|Block), data=data.rcb1, REML=TRUE,
+ control=lmerControl(check.nobs.vs.nRE="ignore"))
> AIC(data.rcb1.lmer, data.rcb1.lmer1) df AIC
data.rcb1.lmer 5 722.1087
data.rcb1.lmer1 10 727.2001> anova(data.rcb1.lmer, data.rcb1.lmer1)Data: data.rcb1
Models:
data.rcb1.lmer: y ~ A + (1 | Block)
data.rcb1.lmer1: y ~ A + (A | Block)
Df AIC BIC logLik deviance Chisq Chi Df Pr(>Chisq)
data.rcb1.lmer 5 729.03 742.30 -359.51 719.03
data.rcb1.lmer1 10 733.98 760.52 -356.99 713.98 5.0529 5 0.4095> summary(data.rcb1.lmer)Linear mixed model fit by REML ['lmerMod']
Formula: y ~ A + (1 | Block)
Data: data.rcb1
Control: lmerControl(check.nobs.vs.nRE = "ignore")
REML criterion at convergence: 712.1
Scaled residuals:
Min 1Q Median 3Q Max
-1.78748 -0.57868 -0.07108 0.49991 2.33728
Random effects:
Groups Name Variance Std.Dev.
Block (Intercept) 132.57 11.514
Residual 20.91 4.572
Number of obs: 105, groups: Block, 35
Fixed effects:
Estimate Std. Error t value
(Intercept) 43.034 2.094 20.55
AB 28.452 1.093 26.03
AC 40.156 1.093 36.74
Correlation of Fixed Effects:
(Intr) AB
AB -0.261
AC -0.261 0.500> anova(data.rcb1.lmer)Analysis of Variance Table
Df Sum Sq Mean Sq F value
A 2 29855 14927 714.03> # Perform SAS-like p-value calculations (requires the lmerTest and pbkrtest package)
> library(lmerTest)
> data.rcb1.lmer <- update(data.rcb1.lmer)
> summary(data.rcb1.lmer)Linear mixed model fit by REML t-tests use Satterthwaite approximations to degrees of freedom [
lmerMod]
Formula: y ~ A + (1 | Block)
Data: data.rcb1
Control: lmerControl(check.nobs.vs.nRE = "ignore")
REML criterion at convergence: 712.1
Scaled residuals:
Min 1Q Median 3Q Max
-1.78748 -0.57868 -0.07108 0.49991 2.33728
Random effects:
Groups Name Variance Std.Dev.
Block (Intercept) 132.57 11.514
Residual 20.91 4.572
Number of obs: 105, groups: Block, 35
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 43.034 2.094 40.930 20.55 <2e-16 ***
AB 28.452 1.093 68.000 26.03 <2e-16 ***
AC 40.156 1.093 68.000 36.74 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation of Fixed Effects:
(Intr) AB
AB -0.261
AC -0.261 0.500> anova(data.rcb1.lmer) # Satterthwaite denominator df methodAnalysis of Variance Table of type III with Satterthwaite
approximation for degrees of freedom
Sum Sq Mean Sq NumDF DenDF F.value Pr(>F)
A 29855 14927 2 68 714.03 < 2.2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1> anova(data.rcb1.lmer,ddf="Kenward-Roger") # Kenward-Roger denominator df methodAnalysis of Variance Table
Df Sum Sq Mean Sq F value
A 2 29855 14927 714.03| Format of tobacco.csv data files | |||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|

|