Multiple Linear Regression
Centering
\(growth = intercept + temperature + nitrogen\)
\(y_i=\beta_0+\beta_1x_{i1}+\beta_2x_{i2}+...+\beta_jx_{ij}+\epsilon_i\)
\(growth = intercept + temperature + nitrogen\)
\(y_i=\beta_0+\beta_1x_{i1}+\beta_2x_{i2}+...+\beta_jx_{ij}+\epsilon_i\)
\(growth = intercept + temp + nitro + temp\times nitro\)
\(y_i=\beta_0+\beta_1x_{i1}+\beta_2x_{i2}+\beta_3x_{i1}x_{i2}+...+\epsilon_i\)
\(growth = intercept + temperature + nitrogen\) \(y_i=\beta_0+\beta_1x_{i1}+\beta_2x_{i2}+...+\beta_jx_{ij}+\epsilon_i\)

Normality, homog., linearity 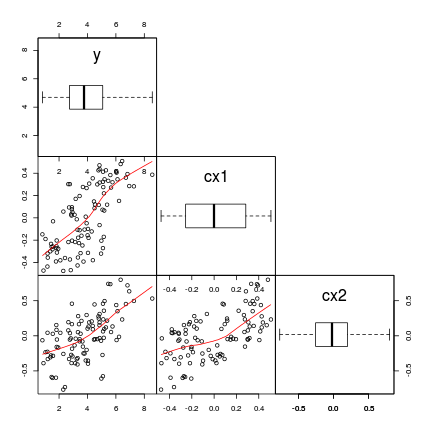
(multi)collinearity 
(multi)collinearity
library(car)
# additive model - scaled predictors
vif(lm(y ~ cx1 + cx2, data)) cx1 cx2
1.744 1.744 (multi)collinearity
library(car)
# additive model - scaled predictors
vif(lm(y ~ cx1 + cx2, data)) cx1 cx2
1.744 1.744 # multiplicative model - raw predictors
vif(lm(y ~ x1 * x2, data)) x1 x2 x1:x2
7.260 5.913 16.949 (multi)collinearity
library(car)
# additive model - scaled predictors
vif(lm(y ~ cx1 + cx2, data)) cx1 cx2
1.744 1.744 # multiplicative model - raw predictors
vif(lm(y ~ x1 * x2, data)) x1 x2 x1:x2
7.260 5.913 16.949 # multiplicative model - scaled predictors
vif(lm(y ~ cx1 * cx2, data)) cx1 cx2 cx1:cx2
1.769 1.772 1.019 loyn <- read.table("../data/loyn.csv", header = T,
sep = ",", strip.white = T)
head(loyn) ABUND AREA YR.ISOL DIST LDIST GRAZE ALT
1 5.3 0.1 1968 39 39 2 160
2 2.0 0.5 1920 234 234 5 60
3 1.5 0.5 1900 104 311 5 140
4 17.1 1.0 1966 66 66 3 160
5 13.8 1.0 1918 246 246 5 140
6 14.1 1.0 1965 234 285 3 130# via car's scatterplotMatrix function
library(car)
scatterplotMatrix(~ABUND + DIST + LDIST + AREA + GRAZE +
ALT + YR.ISOL, data = loyn, diagonal = "boxplot")
scatterplotMatrix(~ABUND + log10(DIST) + log10(LDIST) +
log10(AREA) + GRAZE + ALT + YR.ISOL, data = loyn,
diagonal = "boxplot")
# VIF
loyn.lm <- lm(ABUND ~ log10(DIST) + log10(LDIST) +
log10(AREA) + GRAZE + ALT + YR.ISOL, data = loyn)
vif(loyn.lm) log10(DIST) log10(LDIST) log10(AREA)
1.655 2.010 1.912
GRAZE ALT YR.ISOL
2.525 1.468 1.805 1/vif(loyn.lm) log10(DIST) log10(LDIST) log10(AREA)
0.6044 0.4976 0.5231
GRAZE ALT YR.ISOL
0.3961 0.6812 0.5541
# frequentist lm fit
summary(loyn.lm)
Call:
lm(formula = ABUND ~ log10(DIST) + log10(LDIST) + log10(AREA) +
GRAZE + ALT + YR.ISOL, data = loyn)
Residuals:
Min 1Q Median 3Q Max
-15.651 -2.939 0.529 2.535 15.284
Coefficients:
Estimate Std. Error t value
(Intercept) -125.6972 91.6923 -1.37
log10(DIST) -0.9070 2.6757 -0.34
log10(LDIST) -0.6484 2.1227 -0.31
log10(AREA) 7.4702 1.4649 5.10
GRAZE -1.6677 0.9299 -1.79
ALT 0.0195 0.0240 0.81
YR.ISOL 0.0739 0.0452 1.63
Pr(>|t|)
(Intercept) 0.177
log10(DIST) 0.736
log10(LDIST) 0.761
log10(AREA) 5.5e-06 ***
GRAZE 0.079 .
ALT 0.419
YR.ISOL 0.109
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 6.38 on 49 degrees of freedom
Multiple R-squared: 0.685, Adjusted R-squared: 0.646
F-statistic: 17.8 on 6 and 49 DF, p-value: 8.44e-11
library(MuMIn)
dredge(loyn.lm, rank = "AICc")Global model call: lm(formula = ABUND ~ log10(DIST) + log10(LDIST) + log10(AREA) +
GRAZE + ALT + YR.ISOL, data = loyn)
---
Model selection table
(Int) ALT GRA l10(ARE) l10(DIS)
39 -134.300 -1.902 7.166
40 -141.900 0.02586 -1.601 7.076
7 21.600 -2.854 6.890
55 -113.400 -1.842 7.751
47 -120.500 -1.939 7.487 -2.0480
23 25.740 -2.630 7.709
38 -236.700 0.03623 8.150
8 17.280 0.02468 -2.584 6.799
15 26.630 -2.827 7.298 -2.4750
37 -252.200 8.593
48 -131.800 0.02145 -1.676 7.295 -1.3030
56 -127.700 0.02081 -1.624 7.448
24 22.020 0.01615 -2.502 7.472
53 -223.900 9.213
63 -111.800 -1.884 7.756 -1.2570
31 27.280 -2.671 7.715 -1.2610
16 22.110 0.01850 -2.632 7.126 -1.8480
54 -225.800 0.03206 8.479
46 -233.700 0.03413 8.294 -0.6964
45 -241.900 8.904 -1.8350
64 -125.700 0.01951 -1.668 7.470 -0.9070
61 -224.400 9.235 -0.7046
32 23.560 0.01474 -2.546 7.497 -0.9966
62 -226.000 0.03180 8.492 -0.2380
6 3.959 0.04862 9.062
21 18.670 10.750
22 11.220 0.03555 9.907
14 6.929 0.04490 9.300 -1.1950
5 10.400 9.778
29 18.790 10.760 -0.1055
30 10.670 0.03599 9.879 0.4070
13 16.130 10.200 -2.7660
3 34.370 -4.981
4 28.560 0.03191 -4.597
20 19.190 0.04441 -4.410
19 29.780 -4.958
35 -62.750 -4.440
52 -135.300 0.04913 -3.497
11 32.550 -4.950 0.7777
36 -73.580 0.03285 -4.017
51 -100.100 -4.234
12 23.010 0.03793 -4.448 1.9060
28 19.760 0.04392 -4.429 -0.3745
27 31.190 -4.997 -1.1520
43 -72.190 -4.356 1.1420
44 -95.930 0.04056 -3.742 2.3960
60 -134.700 0.04875 -3.513 -0.2841
59 -98.700 -4.274 -1.1480
50 -389.500 0.08802
58 -386.700 0.08918 1.5880
42 -356.500 0.08167 5.3870
34 -348.500 0.07006
49 -429.000
33 -392.300
41 -402.400 3.5440
57 -428.200 0.5653
18 -8.295 0.11150
10 -8.908 0.10710 5.7560
2 5.598 0.09515
26 -12.280 0.11330 3.2660
1 19.510
9 12.230 3.2870
17 13.900
25 11.390 2.2620
l10(LDI) YR.ISO df logLik AICc delta weight
39 0.07835 5 -180.6 372.3 0.00 0.147
40 0.07991 6 -179.8 373.2 0.93 0.092
7 4 -182.3 373.3 0.99 0.090
55 -1.6460 0.06941 6 -180.0 373.8 1.48 0.070
47 0.07354 6 -180.1 373.9 1.55 0.068
23 -2.1900 5 -181.3 373.9 1.59 0.066
38 0.12480 5 -181.4 374.1 1.75 0.061
8 5 -181.6 374.4 2.05 0.053
15 5 -181.6 374.4 2.06 0.053
37 0.13520 4 -183.0 374.8 2.46 0.043
48 0.07658 7 -179.6 375.5 3.19 0.030
56 -0.9973 0.07419 7 -179.6 375.5 3.22 0.029
24 -1.7160 6 -181.1 375.9 3.59 0.024
53 -1.8930 0.12290 5 -182.4 375.9 3.61 0.024
63 -1.1060 0.06939 7 -179.9 376.1 3.84 0.022
31 -1.6480 6 -181.2 376.2 3.86 0.021
16 6 -181.2 376.2 3.88 0.021
54 -0.8482 0.12050 6 -181.3 376.3 4.04 0.020
46 0.12420 6 -181.4 376.5 4.16 0.018
45 0.13190 5 -182.6 376.5 4.17 0.018
64 -0.6484 0.07387 8 -179.5 378.1 5.82 0.008
61 -1.5930 0.12360 6 -182.3 378.4 6.05 0.007
32 -1.3290 7 -181.0 378.4 6.06 0.007
62 -0.7556 0.12070 7 -181.3 379.0 6.65 0.005
6 4 -187.3 383.5 11.16 0.001
21 -3.6070 4 -187.6 384.0 11.70 0.000
22 -2.4110 5 -186.5 384.3 11.99 0.000
14 5 -187.2 385.7 13.34 0.000
5 3 -189.7 385.8 13.47 0.000
29 -3.5640 5 -187.6 386.4 14.12 0.000
30 -2.5640 6 -186.5 386.8 14.48 0.000
13 4 -189.0 386.8 14.50 0.000
3 3 -194.3 395.1 22.78 0.000
4 4 -193.6 395.9 23.62 0.000
20 2.7440 5 -192.5 396.2 23.91 0.000
19 1.7750 4 -193.8 396.5 24.15 0.000
35 0.04898 4 -193.9 396.6 24.25 0.000
52 3.4700 0.07654 6 -191.5 396.6 24.32 0.000
11 4 -194.3 397.3 25.01 0.000
36 0.05143 5 -193.1 397.4 25.07 0.000
51 2.3020 0.06484 5 -193.1 397.4 25.11 0.000
12 5 -193.3 397.8 25.52 0.000
28 2.8950 6 -192.5 398.7 26.41 0.000
27 2.2720 5 -193.8 398.7 26.44 0.000
43 0.05240 5 -193.8 398.8 26.46 0.000
44 0.05917 6 -192.7 399.1 26.77 0.000
60 3.5830 0.07645 7 -191.5 399.2 26.93 0.000
59 2.7980 0.06482 6 -193.0 399.8 27.50 0.000
50 5.4380 0.19610 5 -197.2 405.7 33.34 0.000
58 4.7540 0.19360 6 -197.1 407.9 35.64 0.000
42 0.18060 5 -198.9 409.0 36.70 0.000
34 0.18350 4 -200.7 410.1 37.82 0.000
49 3.8190 0.22510 4 -202.1 413.0 40.71 0.000
33 0.21120 3 -203.7 413.8 41.53 0.000
41 0.21230 4 -203.0 414.7 42.44 0.000
57 3.5680 0.22430 5 -202.1 415.4 43.10 0.000
18 4.5200 4 -205.5 419.8 47.51 0.000
10 4 -205.8 420.3 48.02 0.000
2 3 -207.4 421.2 48.87 0.000
26 3.1370 5 -205.2 421.5 49.21 0.000
1 2 -211.9 428.0 55.66 0.000
9 3 -211.4 429.3 56.99 0.000
17 2.2060 3 -211.5 429.4 57.11 0.000
25 1.2230 4 -211.3 431.5 59.16 0.000
# 1. Refit most parsimonious model
loyn.lm1 <- lm(ABUND ~ log10(AREA) + GRAZE + YR.ISOL,
data = loyn)
summary(loyn.lm1)
Call:
lm(formula = ABUND ~ log10(AREA) + GRAZE + YR.ISOL, data = loyn)
Residuals:
Min 1Q Median 3Q Max
-14.516 -3.814 0.203 3.127 14.554
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -134.2606 86.3908 -1.55 0.126
log10(AREA) 7.1662 1.2726 5.63 7.3e-07
GRAZE -1.9022 0.8745 -2.18 0.034
YR.ISOL 0.0784 0.0434 1.81 0.077
(Intercept)
log10(AREA) ***
GRAZE *
YR.ISOL .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 6.31 on 52 degrees of freedom
Multiple R-squared: 0.673, Adjusted R-squared: 0.654
F-statistic: 35.7 on 3 and 52 DF, p-value: 1.13e-12# plot(allEffects(loyn.lm2)) 2. model averaging
model.avg(get.models(dredge(loyn.lm, rank = "AICc"),
subset = delta <= 100))
Call:
model.avg.default(object = get.models(dredge(loyn.lm, rank = "AICc"),
subset = delta <= 100))
Component models:
'236' '1236' '23' '2356' '2346'
'235' '136' '123' '234' '36'
'12346' '12356' '1235' '356' '23456'
'2345' '1234' '1356' '1346' '346'
'123456' '3456' '12345' '13456' '13'
'35' '135' '134' '3' '345'
'1345' '34' '2' '12' '125'
'25' '26' '1256' '24' '126'
'256' '124' '1245' '245' '246'
'1246' '12456' '2456' '156' '1456'
'146' '16' '56' '6' '46'
'456' '15' '14' '1' '145'
'(Null)' '4' '5' '45'
Coefficients:
(Intercept) GRAZE log10(AREA)
-98.92575 -2.18001 7.54361
YR.ISOL ALT log10(LDIST)
0.09094 0.02612 -1.59287
log10(DIST)
-1.67918 \(abund = intercept + dist + ldist + area + graze \\+ alt + yr.isol\\\)
\(abund_i \sim{} N(\mu_i, tau)\\ \mu_i = \beta_0+\beta_1 log(dist_{i})+\beta_2 log(ldist_{i})+\beta_3 log(area_{i})\\ +\beta_4 graze_{i} + \beta_5 log(yr.isol_{i})\\\)
\(\beta_{0} \sim{} N(0, 0.0000001)\)
\(\beta_{1} \sim{} N(0, 0.0000001)\)
\(tau = \frac{1}{\sigma^2}\)
\(\sigma \sim{} Gamma(0.001, 0.001)\)
modelString="
model {
#Likelihood
for (i in 1:n) {
abund[i]~dnorm(mu[i],tau)
mu[i] <- beta0 + beta.dist*(dist[i]) + beta.ldist*(ldist[i]) +
beta.area*(area[i]) + beta.graze*(graze[i]) + beta.alt*(alt[i]) +
beta.yr*(yr[i])
y.err[i] <- abund[i] - mu[i]
}
#Priors
beta0 ~ dnorm (0,1.0E-6)
beta.dist ~ dnorm(0,1.0E-6)
beta.ldist ~ dnorm(0,1.0E-6)
beta.area ~ dnorm(0,1.0E-6)
beta.graze ~ dnorm(0,1.0E-6)
beta.alt ~ dnorm(0,1.0E-6)
beta.yr ~ dnorm(0,1.0E-6)
tau <- 1 / (sigma * sigma)
sigma~dgamma(0.001,0.001)
#Other Derived parameters
sd.dist <- abs(beta.dist)*sd(dist[])
sd.ldist <- abs(beta.ldist)*sd(ldist[])
sd.area <- abs(beta.area)*sd(area[])
sd.graze <- abs(beta.graze)*sd(graze[])
sd.alt <- abs(beta.alt)*sd(alt[])
sd.yr <- abs(beta.yr)*sd(yr[])
sd.resid <- sd(y.err)
}
"
writeLines(modelString,con="pres.9.3bQ1.1.bug")
loyn.list <- with(loyn,
list(abund=ABUND,
dist=log10(DIST),
ldist=log10(LDIST),
area=log10(AREA),
graze=GRAZE,
alt=ALT,
yr=YR.ISOL,
n=nrow(loyn)
)
)
#inits <- rep(list(list(alpha=mean(loyn$YIELD), beta=0,
# sigma=sd(loyn$YIELD))),3)
params <- c("beta0","beta.dist","beta.ldist", "beta.area",
"beta.graze","beta.alt","beta.yr",
"sigma","sd.dist","sd.ldist","sd.area",
"sd.graze","sd.alt","sd.yr","sd.resid")
burnInSteps = 2000
nChains = 3
numSavedSteps = 50000
thinSteps = 10
nIter = ceiling((numSavedSteps * thinSteps)/nChains)
library(R2jags)
rnorm(1)[1] -0.5847loyn.r2jags <- jags(data=loyn.list,
inits=NULL,
parameters.to.save=params,
model.file="pres.9.3bQ1.1.bug",
n.chains=3,
n.iter=nIter,
n.burnin=burnInSteps,
n.thin=10
)Compiling model graph
Resolving undeclared variables
Allocating nodes
Graph Size: 708
Initializing modelplot(as.mcmc(loyn.r2jags))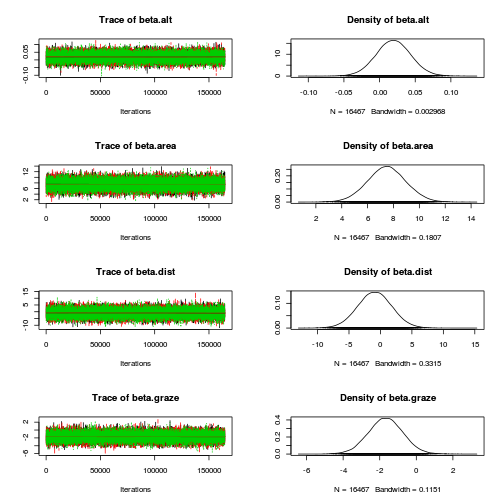
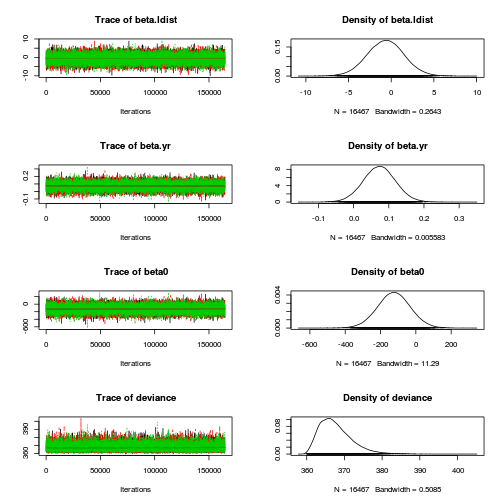
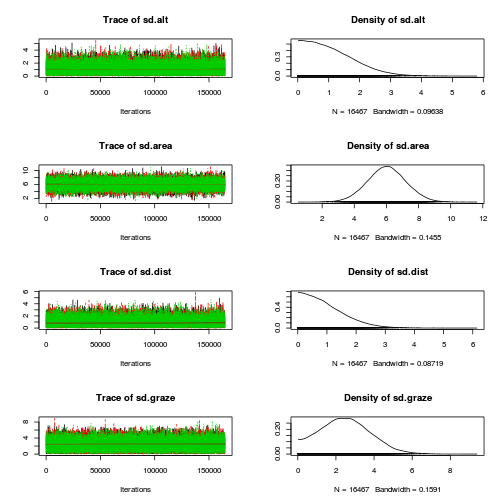
plot(as.mcmc(loyn.r2jags), ask=TRUE)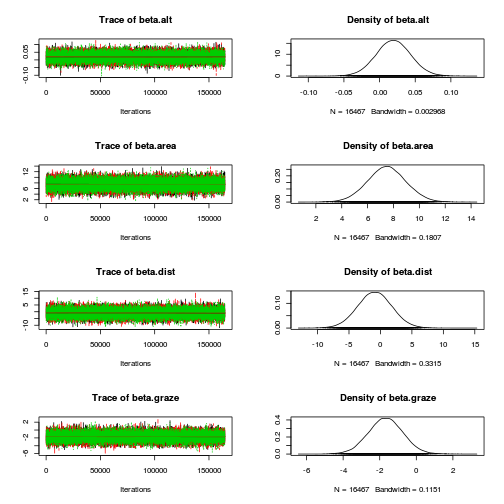
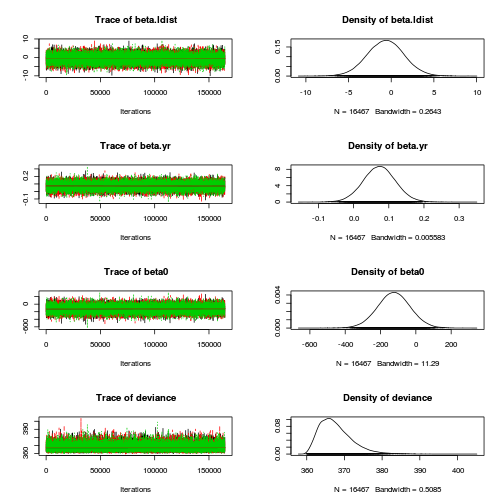
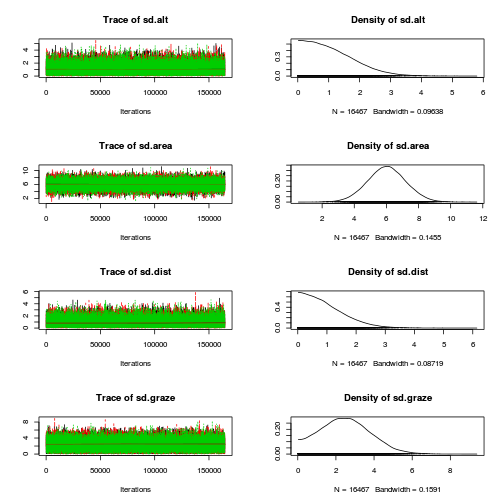
autocorr.diag(as.mcmc(loyn.r2jags)) beta.alt beta.area beta.dist beta.graze
Lag 0 1.000000 1.000000 1.000000 1.000e+00
Lag 10 0.002621 -0.002386 0.003462 3.251e-03
Lag 50 -0.001222 -0.001079 0.001946 -6.453e-05
Lag 100 -0.002387 0.003133 -0.002479 -9.442e-03
Lag 500 0.003377 0.003626 -0.004768 6.225e-03
beta.ldist beta.yr beta0 deviance
Lag 0 1.0000000 1.000000 1.000000 1.0000000
Lag 10 0.0006997 0.006184 0.006598 -0.0002467
Lag 50 -0.0057817 0.003342 0.003025 -0.0040555
Lag 100 0.0028848 0.004238 0.003273 -0.0031883
Lag 500 -0.0052551 -0.002104 -0.002595 0.0012943
sd.alt sd.area sd.dist sd.graze
Lag 0 1.000000 1.000000 1.0000000 1.0000000
Lag 10 0.005823 -0.002386 -0.0003376 0.0032880
Lag 50 -0.003295 -0.001079 0.0013552 0.0003176
Lag 100 -0.003933 0.003133 0.0002549 -0.0089558
Lag 500 0.001510 0.003626 0.0038924 0.0079450
sd.ldist sd.resid sd.yr sigma
Lag 0 1.0000000 1.000000 1.0000000 1.000000
Lag 10 0.0035399 -0.001288 0.0082089 0.003390
Lag 50 -0.0021596 -0.004671 0.0002784 0.007018
Lag 100 -0.0034125 -0.002949 0.0020523 -0.002163
Lag 500 0.0008754 0.003068 -0.0035982 -0.003360
print(loyn.r2jags)Inference for Bugs model at "pres.9.3bQ1.1.bug", fit using jags,
3 chains, each with 166667 iterations (first 2000 discarded), n.thin = 10
n.sims = 49401 iterations saved
mu.vect sd.vect 2.5% 25%
beta.alt 0.019 0.024 -0.029 0.003
beta.area 7.477 1.495 4.546 6.485
beta.dist -0.907 2.732 -6.282 -2.728
beta.graze -1.680 0.952 -3.562 -2.310
beta.ldist -0.653 2.173 -4.931 -2.109
beta.yr 0.073 0.046 -0.017 0.042
beta0 -124.095 93.550 -308.483 -186.023
sd.alt 1.090 0.790 0.045 0.456
sd.area 6.020 1.204 3.660 5.222
sd.dist 0.940 0.714 0.035 0.372
sd.graze 2.495 1.303 0.215 1.537
sd.ldist 1.027 0.785 0.039 0.406
sd.resid 6.336 0.219 6.045 6.176
sd.yr 1.911 1.069 0.134 1.096
sigma 6.473 0.667 5.322 6.003
deviance 367.670 4.322 361.399 364.487
50% 75% 97.5% Rhat n.eff
beta.alt 0.019 0.036 0.068 1.001 37000
beta.area 7.476 8.469 10.415 1.001 49000
beta.dist -0.904 0.911 4.462 1.001 49000
beta.graze -1.679 -1.046 0.179 1.001 47000
beta.ldist -0.646 0.792 3.633 1.001 32000
beta.yr 0.073 0.104 0.164 1.001 49000
beta0 -124.137 -62.120 60.143 1.001 49000
sd.alt 0.950 1.579 2.927 1.001 49000
sd.area 6.020 6.818 8.385 1.001 49000
sd.dist 0.795 1.354 2.662 1.001 49000
sd.graze 2.449 3.367 5.193 1.001 49000
sd.ldist 0.859 1.480 2.923 1.001 49000
sd.resid 6.292 6.448 6.882 1.001 29000
sd.yr 1.858 2.626 4.156 1.001 49000
sigma 6.417 6.880 7.950 1.001 21000
deviance 366.956 370.070 378.010 1.001 18000
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 9.3 and DIC = 377.0
DIC is an estimate of expected predictive error (lower deviance is better).trace<-loyn.r2jags$BUGSoutput$sims.matrix
library(plyr)
apply(trace, 2, mean) beta.alt beta.area beta.dist beta.graze
0.01941 7.47695 -0.90720 -1.68016
beta.ldist beta.yr beta0 deviance
-0.65253 0.07308 -124.09479 367.66997
sd.alt sd.area sd.dist sd.graze
1.09044 6.01988 0.93985 2.49467
sd.ldist sd.resid sd.yr sigma
1.02678 6.33619 1.91134 6.47254 apply(trace, 2, median) beta.alt beta.area beta.dist beta.graze
0.01934 7.47649 -0.90426 -1.67872
beta.ldist beta.yr beta0 deviance
-0.64568 0.07316 -124.13712 366.95643
sd.alt sd.area sd.dist sd.graze
0.94978 6.01951 0.79519 2.44855
sd.ldist sd.resid sd.yr sigma
0.85873 6.29226 1.85811 6.41736
adply(trace, 2, function(x) {
data.frame(Mean=mean(x),Median=median(x), HPDinterval(as.mcmc(x)))
}) X1 Mean Median lower
1 beta.alt 0.01941 0.01934 -2.807e-02
2 beta.area 7.47695 7.47649 4.543e+00
3 beta.dist -0.90720 -0.90426 -6.179e+00
4 beta.graze -1.68016 -1.67872 -3.579e+00
5 beta.ldist -0.65253 -0.64568 -4.896e+00
6 beta.yr 0.07308 0.07316 -1.861e-02
7 beta0 -124.09479 -124.13712 -3.058e+02
8 deviance 367.66997 366.95643 3.606e+02
9 sd.alt 1.09044 0.94978 1.832e-05
10 sd.area 6.01988 6.01951 3.657e+00
11 sd.dist 0.93985 0.79519 4.505e-05
12 sd.graze 2.49467 2.44855 8.624e-05
13 sd.ldist 1.02678 0.85873 2.045e-05
14 sd.resid 6.33619 6.29226 6.005e+00
15 sd.yr 1.91134 1.85811 1.254e-04
16 sigma 6.47254 6.41736 5.197e+00
upper
1 0.06808
2 10.41026
3 4.55262
4 0.16241
5 3.65802
6 0.16264
7 62.77179
8 376.12886
9 2.58078
10 8.38157
11 2.31668
12 4.71880
13 2.53890
14 6.76497
15 3.77027
16 7.77398# just the standard deviations (mean not useful)
adply(trace[,9:15], 2, function(x) {
data.frame(Median=median(x), HPDinterval(as.mcmc(x)))
}) X1 Median lower upper
1 sd.alt 0.9498 1.832e-05 2.581
2 sd.area 6.0195 3.657e+00 8.382
3 sd.dist 0.7952 4.505e-05 2.317
4 sd.graze 2.4486 8.624e-05 4.719
5 sd.ldist 0.8587 2.045e-05 2.539
6 sd.resid 6.2923 6.005e+00 6.765
7 sd.yr 1.8581 1.254e-04 3.770# a more clever way
iCol <- grep("sd", colnames(trace))
sumry <- adply(trace[,iCol], 2, function(x) {
data.frame(Median=median(x), HPDinterval(as.mcmc(x)))
})
sumry$Perc <- 100*sumry$Median/sum(sumry$Median)
sumry X1 Median lower upper Perc
1 sd.alt 0.9498 1.832e-05 2.581 4.941
2 sd.area 6.0195 3.657e+00 8.382 31.316
3 sd.dist 0.7952 4.505e-05 2.317 4.137
4 sd.graze 2.4486 8.624e-05 4.719 12.738
5 sd.ldist 0.8587 2.045e-05 2.539 4.467
6 sd.resid 6.2923 6.005e+00 6.765 32.734
7 sd.yr 1.8581 1.254e-04 3.770 9.667
library(ggplot2)
plt <- ggplot(sumry, aes(y=Median, x=X1))+geom_point()+
geom_errorbar(aes(ymin=lower, ymax=upper), width=0)+
geom_hline(aes(yintercept=0))
plt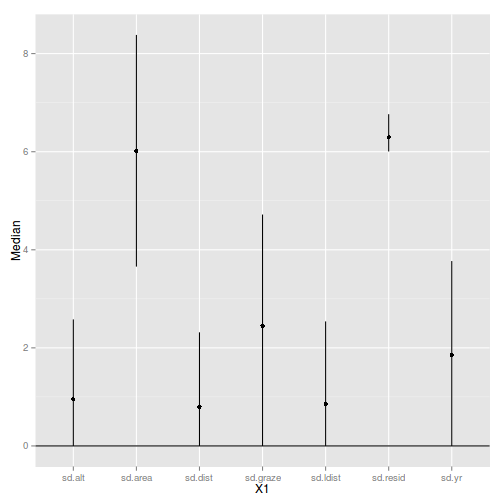
plt+coord_flip()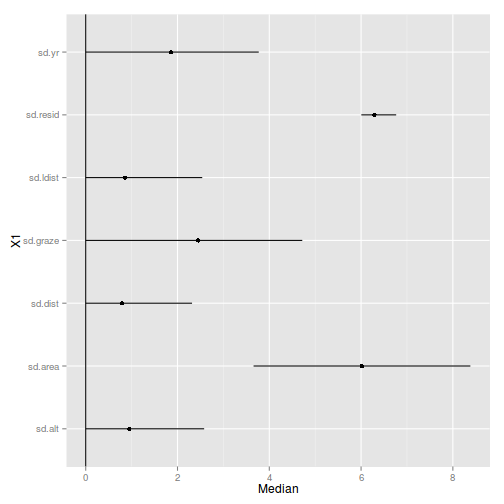
#now lets use the model matrix
modelString="
model {
#Likelihood
for (i in 1:n) {
Y[i]~dnorm(mu[i],tau)
mu[i] <- inprod(beta[],X[i,])
y.err[i] <- Y[i] - mu[i]
}
#Priors
for (i in 1:p) {
beta[i] ~ dnorm (0,1.0E-6)
}
tau <- 1 / (sigma * sigma)
sigma~dgamma(0.001,0.001)
#Other Derived parameters
for (i in 1:p) {
sd[i] <- abs(beta[i])*sd(X[,i])
}
sd.resid <- sd(y.err)
}
"
writeLines(modelString,con="pres.9.3bQ1.2.bug")
Xmat <- model.matrix(~log10(DIST)+
log10(LDIST)+
log10(AREA)+
GRAZE+
ALT+
YR.ISOL,
data=loyn)
loyn.list <- with(loyn,
list(Y=ABUND,
X=Xmat,
p=7,
n=nrow(loyn))
)
#inits <- rep(list(list(alpha=mean(loyn$YIELD), beta=0,
# sigma=sd(loyn$YIELD))),3)
params <- c("beta","sigma","sd","sd.resid")
burnInSteps = 2000
nChains = 3
numSavedSteps = 50000
thinSteps = 10
nIter = ceiling((numSavedSteps * thinSteps)/nChains)
library(R2jags)
loyn.r2jags <- jags(data=loyn.list,
inits=NULL,
parameters.to.save=params,
model.file="pres.9.3bQ1.2.bug",
n.chains=3,
n.iter=nIter,
n.burnin=burnInSteps,
n.thin=10
)Compiling model graph
Resolving undeclared variables
Allocating nodes
Graph Size: 663
Initializing modelprint(loyn.r2jags)Inference for Bugs model at "pres.9.3bQ1.2.bug", fit using jags,
3 chains, each with 166667 iterations (first 2000 discarded), n.thin = 10
n.sims = 49401 iterations saved
mu.vect sd.vect 2.5% 25%
beta[1] -124.245 93.423 -309.005 -186.655
beta[2] -0.925 2.737 -6.327 -2.756
beta[3] -0.648 2.159 -4.913 -2.096
beta[4] 7.473 1.496 4.526 6.480
beta[5] -1.674 0.955 -3.563 -2.313
beta[6] 0.019 0.024 -0.028 0.003
beta[7] 0.073 0.046 -0.017 0.042
sd[1] 0.000 0.000 0.000 0.000
sd[2] 0.944 0.716 0.038 0.377
sd[3] 1.020 0.780 0.041 0.404
sd[4] 6.017 1.204 3.644 5.217
sd[5] 2.487 1.307 0.207 1.521
sd[6] 1.089 0.790 0.043 0.449
sd[7] 1.911 1.073 0.135 1.087
sd.resid 6.338 0.220 6.044 6.178
sigma 6.483 0.667 5.337 6.016
deviance 367.699 4.319 361.458 364.535
50% 75% 97.5% Rhat n.eff
beta[1] -124.031 -61.230 58.141 1.001 49000
beta[2] -0.935 0.900 4.464 1.001 21000
beta[3] -0.640 0.794 3.607 1.001 12000
beta[4] 7.479 8.476 10.408 1.001 17000
beta[5] -1.673 -1.034 0.198 1.001 49000
beta[6] 0.019 0.036 0.068 1.001 49000
beta[7] 0.073 0.104 0.165 1.001 49000
sd[1] 0.000 0.000 0.000 1.000 1
sd[2] 0.797 1.357 2.675 1.001 24000
sd[3] 0.850 1.469 2.922 1.001 18000
sd[4] 6.022 6.824 8.380 1.001 17000
sd[5] 2.441 3.372 5.194 1.001 49000
sd[6] 0.946 1.581 2.920 1.001 30000
sd[7] 1.855 2.634 4.172 1.001 33000
sd.resid 6.294 6.447 6.881 1.001 34000
sigma 6.427 6.893 7.961 1.001 49000
deviance 366.982 370.062 378.025 1.001 48000
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 9.3 and DIC = 377.0
DIC is an estimate of expected predictive error (lower deviance is better).
trace<-loyn.r2jags$BUGSoutput$sims.matrix
iCol <- grep("sd", colnames(trace))
sumry <- adply(trace[,iCol], 2, function(x) {
data.frame(Median=median(x), HPDinterval(as.mcmc(x)))
})
sumry$Perc <- 100*sumry$Median/sum(sumry$Median)
sumry X1 Median lower upper Perc
1 sd[1] 0.0000 0.000e+00 0.000 0.000
2 sd[2] 0.7974 2.399e-05 2.327 4.152
3 sd[3] 0.8502 8.123e-05 2.527 4.427
4 sd[4] 6.0218 3.713e+00 8.438 31.356
5 sd[5] 2.4406 8.364e-04 4.732 12.708
6 sd[6] 0.9457 4.252e-05 2.581 4.924
7 sd[7] 1.8548 6.015e-05 3.784 9.658
8 sd.resid 6.2942 6.009e+00 6.768 32.775
nms <- attr(Xmat,'dimnames')[[2]]
rownames(sumry)<-c(paste("sd.",nms, sep=""),'sd.resid')
sumry X1 Median lower upper
sd.(Intercept) sd[1] 0.0000 0.000e+00 0.000
sd.log10(DIST) sd[2] 0.7974 2.399e-05 2.327
sd.log10(LDIST) sd[3] 0.8502 8.123e-05 2.527
sd.log10(AREA) sd[4] 6.0218 3.713e+00 8.438
sd.GRAZE sd[5] 2.4406 8.364e-04 4.732
sd.ALT sd[6] 0.9457 4.252e-05 2.581
sd.YR.ISOL sd[7] 1.8548 6.015e-05 3.784
sd.resid sd.resid 6.2942 6.009e+00 6.768
Perc
sd.(Intercept) 0.000
sd.log10(DIST) 4.152
sd.log10(LDIST) 4.427
sd.log10(AREA) 31.356
sd.GRAZE 12.708
sd.ALT 4.924
sd.YR.ISOL 9.658
sd.resid 32.775
Xmat <- model.matrix(~scale(log10(DIST))+scale(log10(LDIST))+scale(log10(AREA))+
scale(GRAZE)+scale(ALT)+scale(YR.ISOL), data=loyn)
loyn.list <- with(loyn,
list(Y=ABUND,
X=Xmat,
p=7,
n=nrow(loyn))
)
params <- c("beta","sigma","sd","sd.resid","gamma")
loyn.r2jags <- jags(data=loyn.list,
inits=NULL,
parameters.to.save=params,
model.file="pres.9.3bQ1.2.bug",
n.chains=3,
n.iter=nIter,
n.burnin=burnInSteps,
n.thin=10
)Compiling model graph
Resolving undeclared variables
Allocating nodes
Graph Size: 663
Initializing modelprint(loyn.r2jags)Inference for Bugs model at "pres.9.3bQ1.2.bug", fit using jags,
3 chains, each with 166667 iterations (first 2000 discarded), n.thin = 10
n.sims = 49401 iterations saved
mu.vect sd.vect 2.5% 25% 50%
beta[1] 19.521 0.871 17.809 18.942 19.521
beta[2] -0.377 1.124 -2.575 -1.131 -0.380
beta[3] -0.365 1.237 -2.810 -1.187 -0.367
beta[4] 6.054 1.214 3.654 5.250 6.056
beta[5] -2.460 1.386 -5.207 -3.383 -2.458
beta[6] 0.850 1.066 -1.240 0.139 0.856
beta[7] 1.886 1.182 -0.447 1.098 1.887
sd[1] 0.000 0.000 0.000 0.000 0.000
sd[2] 0.936 0.709 0.038 0.373 0.788
sd[3] 1.015 0.777 0.039 0.403 0.855
sd[4] 6.000 1.203 3.621 5.203 6.002
sd[5] 2.482 1.292 0.209 1.538 2.437
sd[6] 1.097 0.789 0.045 0.462 0.962
sd[7] 1.927 1.074 0.134 1.109 1.873
sd.resid 6.335 0.219 6.044 6.176 6.290
sigma 6.479 0.673 5.337 6.005 6.420
deviance 367.659 4.306 361.408 364.533 366.918
75% 97.5% Rhat n.eff
beta[1] 20.097 21.254 1.001 25000
beta[2] 0.372 1.833 1.001 49000
beta[3] 0.457 2.070 1.001 49000
beta[4] 6.860 8.455 1.001 49000
beta[5] -1.542 0.262 1.001 49000
beta[6] 1.559 2.936 1.001 49000
beta[7] 2.674 4.202 1.001 49000
sd[1] 0.000 0.000 1.000 1
sd[2] 1.349 2.637 1.001 21000
sd[3] 1.461 2.881 1.001 49000
sd[4] 6.798 8.379 1.001 49000
sd[5] 3.353 5.161 1.001 49000
sd[6] 1.582 2.920 1.001 49000
sd[7] 2.651 4.165 1.001 49000
sd.resid 6.445 6.880 1.001 49000
sigma 6.888 7.963 1.001 49000
deviance 370.013 378.032 1.001 49000
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 9.3 and DIC = 376.9
DIC is an estimate of expected predictive error (lower deviance is better).trace<-loyn.r2jags$BUGSoutput$sims.matrix
#Perform Gibbs Variable Selection (Model selection) to determine the most parsimonious model.
modelString="
model {
for (i in 1:p) {
gb[i] <- gamma[i]*beta[i]
}
#Likelihood
for (i in 1:n) {
Y[i]~dnorm(mu[i],tau)
mu[i] <- gamma0*beta0+inprod(X[i,1:p],gb[1:p])
y.err[i] <- Y[i] - mu[i]
}
for (j in 1:p) {
TempIndicator[j] <- gamma[j]*pow(2,j-1)
}
mdl <- 1+sum(TempIndicator[])
for (j in 1:64) {
pmdl[j] <- equals(mdl,j)
}
#Priors
beta0 ~ dnorm (0,1.0E-6)
gamma0 <- 1
for (i in 1:p) {
gamma[i] ~ dbern(0.5)
beta[i] ~ dnorm (bprior[i],tprior[i])
bprior[i] <- (1-gamma0)*mean[i]
tprior[i] <- gamma[i]*0.01+(1-gamma[i])/(se[i]*se[i])
}
tau <- 1 / (sigma * sigma)
sigma~dgamma(0.001,0.001)
#Other Derived parameters
for (i in 1:p) {
sds[i] <- abs(beta[i])*sd(X[,i])
}
sd.resid <- sd(y.err)
}
"
writeLines(modelString,con="pres.9.3bQ1.3.bug")
Xmat <- model.matrix(~-1+scale(log10(DIST))+scale(log10(LDIST))+
scale(log10(AREA))+scale(GRAZE)+scale(ALT)+scale(YR.ISOL), data=loyn)
loyn.betas <- apply(trace[,2:7],2,mean)
loyn.sds <- apply(trace[,2:7],2,sd)
loyn.list <- with(loyn,
list(Y=ABUND,
X=Xmat,
p=6,
n=nrow(loyn),
mean=loyn.betas, se=loyn.sds)
)
#inits <- rep(list(list(alpha=mean(loyn$YIELD), beta=0,
# sigma=sd(loyn$YIELD))),3)
params <- c("beta0","beta","sigma","sds","sd.resid","gamma0","gamma","pmdl")
loyn.r2jags <- jags(data=loyn.list,
inits=NULL,
parameters.to.save=params,
model.file="pres.9.3bQ1.3.bug",
n.chains=3,
n.iter=nIter,
n.burnin=burnInSteps,
n.thin=10
)Compiling model graph
Resolving undeclared variables
Allocating nodes
Graph Size: 871
Initializing modelprint(loyn.r2jags)Inference for Bugs model at "pres.9.3bQ1.3.bug", fit using jags,
3 chains, each with 166667 iterations (first 2000 discarded), n.thin = 10
n.sims = 49401 iterations saved
mu.vect sd.vect 2.5% 25% 50%
beta[1] -0.104 1.138 -2.300 -0.873 -0.111
beta[2] -0.149 1.265 -2.582 -1.013 -0.159
beta[3] 6.024 1.195 3.711 5.217 6.013
beta[4] -2.760 2.046 -5.917 -4.250 -3.170
beta[5] 0.220 1.154 -2.006 -0.559 0.201
beta[6] 1.283 1.785 -1.976 -0.078 1.225
beta0 19.516 0.875 17.784 18.939 19.516
gamma[1] 0.122 0.327 0.000 0.000 0.000
gamma[2] 0.149 0.356 0.000 0.000 0.000
gamma[3] 1.000 0.009 1.000 1.000 1.000
gamma[4] 0.763 0.425 0.000 1.000 1.000
gamma[5] 0.181 0.385 0.000 0.000 0.000
gamma[6] 0.493 0.500 0.000 0.000 0.000
gamma0 1.000 0.000 1.000 1.000 1.000
pmdl[1] 0.000 0.000 0.000 0.000 0.000
pmdl[2] 0.000 0.000 0.000 0.000 0.000
pmdl[3] 0.000 0.000 0.000 0.000 0.000
pmdl[4] 0.000 0.000 0.000 0.000 0.000
pmdl[5] 0.002 0.048 0.000 0.000 0.000
pmdl[6] 0.000 0.021 0.000 0.000 0.000
pmdl[7] 0.002 0.040 0.000 0.000 0.000
pmdl[8] 0.000 0.014 0.000 0.000 0.000
pmdl[9] 0.000 0.006 0.000 0.000 0.000
pmdl[10] 0.000 0.000 0.000 0.000 0.000
pmdl[11] 0.000 0.000 0.000 0.000 0.000
pmdl[12] 0.000 0.000 0.000 0.000 0.000
pmdl[13] 0.305 0.460 0.000 0.000 0.000
pmdl[14] 0.051 0.220 0.000 0.000 0.000
pmdl[15] 0.064 0.245 0.000 0.000 0.000
pmdl[16] 0.008 0.088 0.000 0.000 0.000
pmdl[17] 0.000 0.000 0.000 0.000 0.000
pmdl[18] 0.000 0.000 0.000 0.000 0.000
pmdl[19] 0.000 0.000 0.000 0.000 0.000
pmdl[20] 0.000 0.000 0.000 0.000 0.000
pmdl[21] 0.002 0.045 0.000 0.000 0.000
pmdl[22] 0.000 0.015 0.000 0.000 0.000
pmdl[23] 0.000 0.018 0.000 0.000 0.000
pmdl[24] 0.000 0.006 0.000 0.000 0.000
pmdl[25] 0.000 0.000 0.000 0.000 0.000
pmdl[26] 0.000 0.000 0.000 0.000 0.000
pmdl[27] 0.000 0.004 0.000 0.000 0.000
pmdl[28] 0.000 0.000 0.000 0.000 0.000
pmdl[29] 0.055 0.228 0.000 0.000 0.000
pmdl[30] 0.007 0.084 0.000 0.000 0.000
pmdl[31] 0.009 0.096 0.000 0.000 0.000
pmdl[32] 0.001 0.033 0.000 0.000 0.000
pmdl[33] 0.000 0.000 0.000 0.000 0.000
pmdl[34] 0.000 0.000 0.000 0.000 0.000
pmdl[35] 0.000 0.000 0.000 0.000 0.000
pmdl[36] 0.000 0.000 0.000 0.000 0.000
pmdl[37] 0.126 0.332 0.000 0.000 0.000
pmdl[38] 0.016 0.126 0.000 0.000 0.000
pmdl[39] 0.022 0.148 0.000 0.000 0.000
pmdl[40] 0.002 0.047 0.000 0.000 0.000
pmdl[41] 0.000 0.000 0.000 0.000 0.000
pmdl[42] 0.000 0.000 0.000 0.000 0.000
pmdl[43] 0.000 0.004 0.000 0.000 0.000
pmdl[44] 0.000 0.000 0.000 0.000 0.000
pmdl[45] 0.168 0.374 0.000 0.000 0.000
pmdl[46] 0.022 0.147 0.000 0.000 0.000
pmdl[47] 0.026 0.158 0.000 0.000 0.000
pmdl[48] 0.003 0.057 0.000 0.000 0.000
pmdl[49] 0.000 0.000 0.000 0.000 0.000
pmdl[50] 0.000 0.000 0.000 0.000 0.000
pmdl[51] 0.000 0.000 0.000 0.000 0.000
pmdl[52] 0.000 0.000 0.000 0.000 0.000
pmdl[53] 0.051 0.221 0.000 0.000 0.000
pmdl[54] 0.005 0.072 0.000 0.000 0.000
pmdl[55] 0.006 0.075 0.000 0.000 0.000
pmdl[56] 0.001 0.026 0.000 0.000 0.000
pmdl[57] 0.000 0.000 0.000 0.000 0.000
pmdl[58] 0.000 0.000 0.000 0.000 0.000
pmdl[59] 0.000 0.000 0.000 0.000 0.000
pmdl[60] 0.000 0.000 0.000 0.000 0.000
pmdl[61] 0.035 0.184 0.000 0.000 0.000
pmdl[62] 0.004 0.061 0.000 0.000 0.000
pmdl[63] 0.004 0.064 0.000 0.000 0.000
pmdl[64] 0.000 0.022 0.000 0.000 0.000
sd.resid 6.367 0.171 6.101 6.270 6.346
sds[1] 0.903 0.683 0.035 0.358 0.761
sds[2] 1.011 0.757 0.039 0.405 0.858
sds[3] 5.970 1.183 3.677 5.170 5.959
sds[4] 3.003 1.605 0.160 1.711 3.155
sds[5] 0.925 0.706 0.034 0.367 0.776
sds[6] 1.759 1.286 0.066 0.668 1.498
sigma 6.517 0.657 5.381 6.052 6.466
deviance 368.248 3.585 362.532 365.862 367.730
75% 97.5% Rhat n.eff
beta[1] 0.653 2.142 1.001 49000
beta[2] 0.708 2.347 1.001 18000
beta[3] 6.830 8.357 1.001 22000
beta[4] -1.481 1.755 1.001 45000
beta[5] 0.986 2.525 1.001 49000
beta[6] 2.680 4.565 1.001 48000
beta0 20.100 21.236 1.001 49000
gamma[1] 0.000 1.000 1.001 49000
gamma[2] 0.000 1.000 1.001 49000
gamma[3] 1.000 1.000 1.030 49000
gamma[4] 1.000 1.000 1.001 49000
gamma[5] 0.000 1.000 1.001 19000
gamma[6] 1.000 1.000 1.001 49000
gamma0 1.000 1.000 1.000 1
pmdl[1] 0.000 0.000 1.000 1
pmdl[2] 0.000 0.000 1.000 1
pmdl[3] 0.000 0.000 1.000 1
pmdl[4] 0.000 0.000 1.000 1
pmdl[5] 0.000 0.000 1.001 49000
pmdl[6] 0.000 0.000 1.041 27000
pmdl[7] 0.000 0.000 1.002 49000
pmdl[8] 0.000 0.000 1.005 49000
pmdl[9] 0.000 0.000 1.106 49000
pmdl[10] 0.000 0.000 1.000 1
pmdl[11] 0.000 0.000 1.000 1
pmdl[12] 0.000 0.000 1.000 1
pmdl[13] 1.000 1.000 1.001 21000
pmdl[14] 0.000 1.000 1.001 49000
pmdl[15] 0.000 1.000 1.001 49000
pmdl[16] 0.000 0.000 1.003 22000
pmdl[17] 0.000 0.000 1.000 1
pmdl[18] 0.000 0.000 1.000 1
pmdl[19] 0.000 0.000 1.000 1
pmdl[20] 0.000 0.000 1.000 1
pmdl[21] 0.000 0.000 1.004 49000
pmdl[22] 0.000 0.000 1.028 49000
pmdl[23] 0.000 0.000 1.065 21000
pmdl[24] 0.000 0.000 1.291 25000
pmdl[25] 0.000 0.000 1.000 1
pmdl[26] 0.000 0.000 1.000 1
pmdl[27] 0.000 0.000 1.291 49000
pmdl[28] 0.000 0.000 1.000 1
pmdl[29] 0.000 1.000 1.001 8200
pmdl[30] 0.000 0.000 1.001 49000
pmdl[31] 0.000 0.000 1.001 49000
pmdl[32] 0.000 0.000 1.004 49000
pmdl[33] 0.000 0.000 1.000 1
pmdl[34] 0.000 0.000 1.000 1
pmdl[35] 0.000 0.000 1.000 1
pmdl[36] 0.000 0.000 1.000 1
pmdl[37] 0.000 1.000 1.001 49000
pmdl[38] 0.000 0.000 1.001 33000
pmdl[39] 0.000 0.000 1.001 41000
pmdl[40] 0.000 0.000 1.002 49000
pmdl[41] 0.000 0.000 1.000 1
pmdl[42] 0.000 0.000 1.000 1
pmdl[43] 0.000 0.000 1.291 49000
pmdl[44] 0.000 0.000 1.000 1
pmdl[45] 0.000 1.000 1.001 49000
pmdl[46] 0.000 0.000 1.001 49000
pmdl[47] 0.000 1.000 1.001 15000
pmdl[48] 0.000 0.000 1.001 49000
pmdl[49] 0.000 0.000 1.000 1
pmdl[50] 0.000 0.000 1.000 1
pmdl[51] 0.000 0.000 1.000 1
pmdl[52] 0.000 0.000 1.000 1
pmdl[53] 0.000 1.000 1.001 49000
pmdl[54] 0.000 0.000 1.001 49000
pmdl[55] 0.000 0.000 1.004 23000
pmdl[56] 0.000 0.000 1.012 49000
pmdl[57] 0.000 0.000 1.000 1
pmdl[58] 0.000 0.000 1.000 1
pmdl[59] 0.000 0.000 1.000 1
pmdl[60] 0.000 0.000 1.000 1
pmdl[61] 0.000 1.000 1.001 49000
pmdl[62] 0.000 0.000 1.012 11000
pmdl[63] 0.000 0.000 1.003 47000
pmdl[64] 0.000 0.000 1.003 49000
sd.resid 6.442 6.790 1.001 49000
sds[1] 1.306 2.544 1.001 49000
sds[2] 1.465 2.804 1.001 49000
sds[3] 6.769 8.282 1.001 25000
sds[4] 4.213 5.864 1.001 35000
sds[5] 1.336 2.620 1.001 49000
sds[6] 2.684 4.524 1.001 49000
sigma 6.927 7.942 1.001 49000
deviance 370.090 376.925 1.001 49000
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 6.4 and DIC = 374.7
DIC is an estimate of expected predictive error (lower deviance is better).
iCol <- grep("pmdl.*",colnames(as.mcmc(loyn.r2jags$BUGSoutput$sims.matrix)))
loyn.mcmc<-as.mcmc(loyn.r2jags$BUGSoutput$sims.matrix)[,iCol]
#https://docs.google.com/a/monash.edu/viewer?a=v&q=cache:oBkmBL0jEoAJ:homepage.stat.uiowa.edu/~kcowles/s138_2000/GVSpresentation.ppt+bayesian+model+selection+BUGS&hl=en&gl=au&pid=bl&srcid=ADGEESioTEwqpoWMn6VvFVCrp6WuVlv5oVG6Ay5LElylXxx5VOq3vJUpimBIdQWWlyWWfRAXBrbXoPmTpu-LGZNKtuHc2lKWCcBf1h9DUifdSprwP91UGDz2dKixCkLOT35G_sNUAhal&sig=AHIEtbSGjiGN59rRECwpnemhtXxLBRWbnQ
loyn.sum <- apply(loyn.mcmc,2,mean)
ind<-function(p){
if(p == 0) {
return(t <- 0)
} else if(p == 1) {
return(t <- rbind(0, 1))
} else if(p == 2) {
return(t <- rbind(c(0, 0), c(1, 0), c(0, 1), c(1, 1)))
} else {
t <- rbind(cbind(ind(p - 1), rep(0, 2^(p - 1))),
cbind(ind(p - 1), rep(1, 2^(p - 1))))
return(t)
}
}
print.ind<-function(p) {
t <- ind(p)
print("intercept")
ee <-NULL
for(i in 2:nrow(t)) {
e <- NULL
L <- T
for(j in 1:ncol(t)) {
if(t[i, j] == 1 & L == T) {
e <- paste(e, "x", j, sep = "")
L <- F
} else if(t[i, j] == 1 & L == F) {
e <- paste(e, " + x", j, sep = "")
}
}
ee[i]<-e
}
ee
}
ms<-data.frame(print.ind(6), sum=round(loyn.sum,3))[1] "intercept"ms[rev(order(ms$sum)),] print.ind.6. sum
pmdl[13] x3 + x4 0.305
pmdl[45] x3 + x4 + x6 0.168
pmdl[37] x3 + x6 0.126
pmdl[15] x2 + x3 + x4 0.064
pmdl[29] x3 + x4 + x5 0.055
pmdl[53] x3 + x5 + x6 0.051
pmdl[14] x1 + x3 + x4 0.051
pmdl[61] x3 + x4 + x5 + x6 0.035
pmdl[47] x2 + x3 + x4 + x6 0.026
pmdl[46] x1 + x3 + x4 + x6 0.022
pmdl[39] x2 + x3 + x6 0.022
pmdl[38] x1 + x3 + x6 0.016
pmdl[31] x2 + x3 + x4 + x5 0.009
pmdl[16] x1 + x2 + x3 + x4 0.008
pmdl[30] x1 + x3 + x4 + x5 0.007
pmdl[55] x2 + x3 + x5 + x6 0.006
pmdl[54] x1 + x3 + x5 + x6 0.005
pmdl[63] x2 + x3 + x4 + x5 + x6 0.004
pmdl[62] x1 + x3 + x4 + x5 + x6 0.004
pmdl[48] x1 + x2 + x3 + x4 + x6 0.003
pmdl[40] x1 + x2 + x3 + x6 0.002
pmdl[21] x3 + x5 0.002
pmdl[7] x2 + x3 0.002
pmdl[5] x3 0.002
pmdl[56] x1 + x2 + x3 + x5 + x6 0.001
pmdl[32] x1 + x2 + x3 + x4 + x5 0.001
pmdl[64] x1 + x2 + x3 + x4 + x5 + x6 0.000
pmdl[60] x1 + x2 + x4 + x5 + x6 0.000
pmdl[59] x2 + x4 + x5 + x6 0.000
pmdl[58] x1 + x4 + x5 + x6 0.000
pmdl[57] x4 + x5 + x6 0.000
pmdl[52] x1 + x2 + x5 + x6 0.000
pmdl[51] x2 + x5 + x6 0.000
pmdl[50] x1 + x5 + x6 0.000
pmdl[49] x5 + x6 0.000
pmdl[44] x1 + x2 + x4 + x6 0.000
pmdl[43] x2 + x4 + x6 0.000
pmdl[42] x1 + x4 + x6 0.000
pmdl[41] x4 + x6 0.000
pmdl[36] x1 + x2 + x6 0.000
pmdl[35] x2 + x6 0.000
pmdl[34] x1 + x6 0.000
pmdl[33] x6 0.000
pmdl[28] x1 + x2 + x4 + x5 0.000
pmdl[27] x2 + x4 + x5 0.000
pmdl[26] x1 + x4 + x5 0.000
pmdl[25] x4 + x5 0.000
pmdl[24] x1 + x2 + x3 + x5 0.000
pmdl[23] x2 + x3 + x5 0.000
pmdl[22] x1 + x3 + x5 0.000
pmdl[20] x1 + x2 + x5 0.000
pmdl[19] x2 + x5 0.000
pmdl[18] x1 + x5 0.000
pmdl[17] x5 0.000
pmdl[12] x1 + x2 + x4 0.000
pmdl[11] x2 + x4 0.000
pmdl[10] x1 + x4 0.000
pmdl[9] x4 0.000
pmdl[8] x1 + x2 + x3 0.000
pmdl[6] x1 + x3 0.000
pmdl[4] x1 + x2 0.000
pmdl[3] x2 0.000
pmdl[2] x1 0.000
pmdl[1] <NA> 0.000paruelo <- read.table("../data/paruelo.csv", header = T,
sep = ",", strip.white = T)
head(paruelo) C3 LAT LONG MAP MAT JJAMAP DJFMAP
1 0.65 46.40 119.55 199 12.4 0.12 0.45
2 0.65 47.32 114.27 469 7.5 0.24 0.29
3 0.76 45.78 110.78 536 7.2 0.24 0.20
4 0.75 43.95 101.87 476 8.2 0.35 0.15
5 0.33 46.90 102.82 484 4.8 0.40 0.14
6 0.03 38.87 99.38 623 12.0 0.40 0.11# via car's scatterplotMatrix function
library(car)
scatterplotMatrix(~C3 + LAT + LONG + MAP + MAT + JJAMAP +
DJFMAP, data = paruelo, diagonal = "boxplot")
# via lattice
library(lattice)
splom.lat <- splom(paruelo, type = c("p", "r"))
print(splom.lat)
# via ggplot2 - warning these are slow!
library(GGally)
ggpairs(paruelo, lower = list(continuous = "smooth"),
diag = list(continuous = "density"), axisLabels = "none")
# library(ggplot2) splom.gg <-
# plotmatrix(paruelo)+geom_smooth(method='lm')
# print(splom.gg)
# via car's scatterplotMatrix function
library(car)
scatterplotMatrix(~sqrt(C3) + LAT + LONG + MAP + MAT +
JJAMAP + log10(DJFMAP), data = paruelo, diagonal = "boxplot")
cor(paruelo[, -1]) LAT LONG MAP MAT
LAT 1.00000 0.09655 -0.2465 -0.838590
LONG 0.09655 1.00000 -0.7337 -0.213109
MAP -0.24651 -0.73369 1.0000 0.355091
MAT -0.83859 -0.21311 0.3551 1.000000
JJAMAP 0.07417 -0.49156 0.1123 -0.080771
DJFMAP -0.06512 0.77074 -0.4045 0.001478
JJAMAP DJFMAP
LAT 0.07417 -0.065125
LONG -0.49156 0.770744
MAP 0.11226 -0.404512
MAT -0.08077 0.001478
JJAMAP 1.00000 -0.791540
DJFMAP -0.79154 1.000000
# VIF
library(car)
vif(lm(sqrt(paruelo$C3) ~ LAT + LONG + MAP + MAT +
JJAMAP + log10(DJFMAP), data = paruelo)) LAT LONG MAP
3.560 4.988 2.794
MAT JJAMAP log10(DJFMAP)
3.752 3.195 5.467 library(car)
1/vif(lm(sqrt(paruelo$C3) ~ LAT + LONG + MAP + MAT +
JJAMAP + log10(DJFMAP), data = paruelo)) LAT LONG MAP
0.2809 0.2005 0.3579
MAT JJAMAP log10(DJFMAP)
0.2665 0.3130 0.1829
library(car)
scatterplotMatrix(~sqrt(C3) + LAT + LONG, data = paruelo,
diagonal = "boxplot")
vif(lm(sqrt(C3) ~ LAT + LONG, data = paruelo)) LAT LONG
1.009 1.009 vif(lm(sqrt(C3) ~ LAT * LONG, data = paruelo)) LAT LONG LAT:LONG
307.74 66.78 400.94
paruelo$cLAT <- scale(paruelo$LAT, scale = F)
paruelo$cLONG <- scale(paruelo$LONG, scale = F)
vif(lm(sqrt(C3) ~ cLAT * cLONG, data = paruelo)) cLAT cLONG cLAT:cLONG
1.209 1.021 1.220
paruelo.lm <- lm(sqrt(C3) ~ cLAT * cLONG, data = paruelo)
# setup a 2x6 plotting device with small margins
par(mfrow = c(2, 3), oma = c(0, 0, 2, 0))
plot(paruelo.lm, ask = F, which = 1:6)
influence.measures(paruelo.lm)Influence measures of
lm(formula = sqrt(C3) ~ cLAT * cLONG, data = paruelo) :
dfb.1_ dfb.cLAT dfb.cLON dfb.cLAT.
1 -0.02402 -0.083059 -0.084079 -0.121467
2 -0.02221 -0.064489 -0.040666 -0.073805
3 0.08130 0.134833 0.066766 0.117108
4 0.18842 0.102837 -0.149242 -0.074613
5 -0.06669 -0.070090 0.046655 0.023640
6 -0.14663 0.020076 0.158421 0.006884
7 -0.06266 0.095315 -0.006186 0.047476
8 -0.14641 0.034868 0.221772 -0.071379
9 -0.03667 0.026450 0.024624 -0.005908
10 -0.14355 -0.014721 0.043712 0.016311
11 -0.06802 -0.077774 0.052091 0.030960
12 -0.09834 0.126501 0.005986 0.046997
13 0.11115 -0.093743 -0.111372 0.092157
14 0.08474 0.046585 -0.103485 -0.077405
15 -0.10466 0.005614 0.159085 0.003719
16 0.02997 0.006031 -0.022676 -0.007923
17 -0.22231 -0.242042 -0.256005 -0.259471
18 -0.15718 -0.195191 -0.167613 -0.197664
19 -0.03827 0.067695 0.013203 -0.004632
20 0.14881 0.082543 -0.079749 -0.027279
21 0.02694 -0.052154 0.013621 -0.042509
22 0.01333 0.036162 0.002225 0.019532
23 0.03874 -0.041008 -0.026165 0.016163
24 0.18953 -0.068780 -0.263543 0.120259
25 -0.30655 -0.141051 -0.348442 -0.174237
26 -0.01163 0.020739 -0.004288 0.014771
27 -0.06640 -0.035776 0.047618 0.021757
28 0.15820 -0.100195 0.156872 -0.087023
29 0.23271 -0.199428 0.178733 -0.171942
30 -0.05886 -0.051722 0.023280 0.001329
31 0.18206 -0.039000 0.294547 -0.009475
32 0.04158 0.010092 0.004277 0.002215
33 -0.11614 0.057069 -0.084317 0.045564
34 0.17140 -0.010972 0.149470 0.000787
35 0.16157 -0.063790 0.207065 -0.047488
36 -0.16599 0.051605 0.046986 0.028324
37 -0.09520 0.123426 -0.073860 0.112439
38 -0.12248 0.136168 0.016438 0.044325
39 0.21589 0.005332 0.135038 0.008527
40 -0.15732 -0.003358 -0.079732 -0.002338
41 -0.08534 0.010451 -0.044041 0.007825
42 -0.03339 0.004442 -0.028421 0.002067
43 0.11153 -0.008546 0.093501 -0.001341
44 0.06621 0.080305 -0.003529 0.027570
45 0.14322 0.175378 -0.007707 0.060327
46 0.00242 0.002970 -0.000135 0.001018
47 0.04762 0.057578 -0.003518 0.018818
48 0.09643 0.105320 -0.013197 0.027624
49 0.21782 0.063120 -0.324557 -0.197238
50 -0.00148 0.001425 0.002076 -0.003405
51 -0.00449 0.005425 0.006061 -0.012462
52 0.02355 -0.011871 -0.037630 0.035697
53 -0.00616 0.005595 0.008294 -0.011850
54 0.11316 -0.020147 0.059511 -0.015058
55 0.08401 -0.006408 0.050234 -0.003848
56 0.01263 0.000401 0.008831 0.000749
57 0.23558 0.012600 0.162103 0.017720
58 0.15162 0.005056 0.085545 0.005265
59 -0.10917 0.059550 -0.109799 0.050149
60 -0.01928 0.010518 -0.019393 0.008857
61 0.07137 -0.085274 -0.078631 0.125075
62 -0.05053 0.001789 -0.075856 -0.007046
63 0.00374 0.000609 -0.000752 -0.000272
64 -0.03243 -0.005963 0.008964 0.002938
65 -0.17360 -0.059882 0.041757 0.006784
66 -0.21254 -0.161201 0.312816 0.383538
67 0.05944 0.032095 -0.092820 -0.099439
68 -0.03086 0.033212 -0.039691 0.034038
69 -0.08462 0.114424 -0.120547 0.127015
70 -0.12444 0.139820 -0.161230 0.144966
71 -0.09511 0.010274 0.128949 -0.001656
72 0.01563 0.003182 -0.030673 -0.027216
73 -0.06408 -0.180207 0.005698 -0.072658
dffit cov.r cook.d hat inf
1 -0.15232 1.379 5.88e-03 0.2347 *
2 -0.09560 1.227 2.32e-03 0.1389 *
3 0.18920 1.083 9.00e-03 0.0556
4 0.27640 0.957 1.87e-02 0.0317
5 -0.11683 1.091 3.45e-03 0.0448
6 -0.21885 1.001 1.19e-02 0.0305
7 -0.11206 1.102 3.17e-03 0.0510
8 -0.30117 1.016 2.25e-02 0.0520
9 -0.05703 1.088 8.24e-04 0.0317
10 -0.15090 0.989 5.65e-03 0.0153
11 -0.12872 1.101 4.19e-03 0.0534
12 -0.15816 1.064 6.29e-03 0.0388
13 0.24977 1.061 1.56e-02 0.0580
14 0.16432 1.104 6.81e-03 0.0622
15 -0.19200 1.063 9.25e-03 0.0460
16 0.03832 1.082 3.72e-04 0.0234
17 -0.46135 0.867 5.07e-02 0.0464
18 -0.33555 0.980 2.77e-02 0.0483
19 -0.08826 1.133 1.97e-03 0.0703
20 0.19334 0.993 9.27e-03 0.0238
21 0.06315 1.184 1.01e-03 0.1064 *
22 0.03980 1.165 4.02e-04 0.0911
23 0.07567 1.106 1.45e-03 0.0475
24 0.39649 0.949 3.83e-02 0.0522
25 -0.50904 0.721 5.92e-02 0.0333 *
26 -0.02450 1.151 1.52e-04 0.0797
27 -0.09331 1.073 2.20e-03 0.0287
28 0.24687 1.004 1.51e-02 0.0371
29 0.36130 0.913 3.16e-02 0.0383
30 -0.08413 1.075 1.79e-03 0.0278
31 0.34876 0.977 2.98e-02 0.0503
32 0.04358 1.068 4.81e-04 0.0147
33 -0.15421 1.033 5.95e-03 0.0260
34 0.22867 0.961 1.29e-02 0.0241
35 0.27074 0.999 1.81e-02 0.0405
36 -0.17882 0.964 7.89e-03 0.0163
37 -0.18180 1.101 8.32e-03 0.0642
38 -0.18258 1.034 8.33e-03 0.0325
39 0.25689 0.889 1.59e-02 0.0190
40 -0.17776 0.972 7.81e-03 0.0172
41 -0.09659 1.047 2.35e-03 0.0177
42 -0.04413 1.081 4.93e-04 0.0240
43 0.14632 1.030 5.36e-03 0.0234
44 0.10631 1.074 2.85e-03 0.0321
45 0.23128 0.999 1.33e-02 0.0324
46 0.00392 1.096 3.89e-06 0.0325
47 0.07632 1.084 1.47e-03 0.0321
48 0.14620 1.048 5.37e-03 0.0294
49 0.43369 0.971 4.59e-02 0.0654
50 -0.00566 1.218 8.12e-06 0.1296 *
51 -0.01985 1.262 1.00e-04 0.1598 *
52 0.06961 1.160 1.23e-03 0.0885
53 -0.02099 1.190 1.12e-04 0.1093 *
54 0.12914 1.024 4.18e-03 0.0181
55 0.09836 1.049 2.44e-03 0.0188
56 0.01556 1.081 6.14e-05 0.0203
57 0.28935 0.857 2.00e-02 0.0201
58 0.17570 0.979 7.64e-03 0.0181
59 -0.16684 1.050 6.98e-03 0.0349
60 -0.02947 1.097 2.20e-04 0.0349
61 0.23769 1.156 1.42e-02 0.1077
62 -0.09174 1.096 2.13e-03 0.0434
63 0.00388 1.076 3.83e-06 0.0148
64 -0.03431 1.072 2.98e-04 0.0155
65 -0.19020 0.951 8.89e-03 0.0164
66 -0.59693 1.136 8.80e-02 0.1617
67 0.15603 1.217 6.16e-03 0.1367 *
68 -0.06400 1.142 1.04e-03 0.0743
69 -0.20622 1.172 1.07e-02 0.1129
70 -0.26402 1.097 1.75e-02 0.0789
71 -0.16322 1.063 6.70e-03 0.0398
72 0.04310 1.251 4.71e-04 0.1534 *
73 -0.19407 1.154 9.51e-03 0.0995
summary(paruelo.lm)
Call:
lm(formula = sqrt(C3) ~ cLAT * cLONG, data = paruelo)
Residuals:
Min 1Q Median 3Q Max
-0.5131 -0.1343 -0.0113 0.1409 0.3894
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.428266 0.023435 18.27 < 2e-16
cLAT 0.043694 0.004867 8.98 3.3e-13
cLONG -0.002877 0.003684 -0.78 0.4375
cLAT:cLONG 0.002282 0.000747 3.06 0.0032
(Intercept) ***
cLAT ***
cLONG
cLAT:cLONG **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.199 on 69 degrees of freedom
Multiple R-squared: 0.54, Adjusted R-squared: 0.52
F-statistic: 27 on 3 and 69 DF, p-value: 1.13e-11
# via the car package
avPlots(paruelo.lm, ask = F, indentify.points = F)
# via the effects package
library(effects)
plot(allEffects(paruelo.lm, default.levels = 5), ask = F)
plot(allEffects(paruelo.lm, default.levels = 10), ask = F)
# 2 standard deviations below the mean longitude
(LongM2 <- mean(paruelo$cLONG) - 2 * sd(paruelo$cLONG))[1] -12.87# 1 standard deviation below the mean longitude
(LongM1 <- mean(paruelo$cLONG) - 1 * sd(paruelo$cLONG))[1] -6.435# mean longitude
(LongM <- mean(paruelo$cLONG))[1] -6.424e-15# 1 standard deviation above the mean longitude
(LongP1 <- mean(paruelo$cLONG) + 1 * sd(paruelo$cLONG))[1] 6.435# 2 standard deviation below the mean longitude
(LongP2 <- mean(paruelo$cLONG) + 2 * sd(paruelo$cLONG))[1] 12.87
(LatM2 <- summary(paruelo.lmM2 <- lm(sqrt(C3) ~ cLAT *
c(cLONG + LongM2), data = paruelo))$coef["cLAT",
]) Estimate Std. Error t value Pr(>|t|)
7.307e-02 1.242e-02 5.884e+00 1.302e-07 (LatM1 <- summary(paruelo.lmM2 <- lm(sqrt(C3) ~ cLAT *
c(cLONG + LongM1), data = paruelo))$coef["cLAT",
]) Estimate Std. Error t value Pr(>|t|)
5.838e-02 8.114e-03 7.195e+00 5.877e-10 (LatM <- summary(paruelo.lmM2 <- lm(sqrt(C3) ~ cLAT *
c(cLONG + LongM), data = paruelo))$coef["cLAT",
]) Estimate Std. Error t value Pr(>|t|)
4.369e-02 4.867e-03 8.977e+00 3.279e-13 (LatP1 <- summary(paruelo.lmM2 <- lm(sqrt(C3) ~ cLAT *
c(cLONG + LongP1), data = paruelo))$coef["cLAT",
]) Estimate Std. Error t value Pr(>|t|)
2.901e-02 5.270e-03 5.504e+00 5.926e-07 (LatP2 <- summary(paruelo.lmM2 <- lm(sqrt(C3) ~ cLAT *
c(cLONG + LongP2), data = paruelo))$coef["cLAT",
]) Estimate Std. Error t value Pr(>|t|)
0.014317 0.008837 1.620090 0.109775 rbind(`Lattitude at Long-2sd` = LatM2, `Lattitude at Long-1sd` = LatM1,
`Lattitude at mean Long` = LatM, `Lattitude at Long+1sd` = LatP1,
`Lattitude at Long+2sd` = LatP2) Estimate Std. Error
Lattitude at Long-2sd 0.07307 0.012418
Lattitude at Long-1sd 0.05838 0.008114
Lattitude at mean Long 0.04369 0.004867
Lattitude at Long+1sd 0.02901 0.005270
Lattitude at Long+2sd 0.01432 0.008837
t value Pr(>|t|)
Lattitude at Long-2sd 5.884 1.302e-07
Lattitude at Long-1sd 7.195 5.877e-10
Lattitude at mean Long 8.977 3.279e-13
Lattitude at Long+1sd 5.504 5.926e-07
Lattitude at Long+2sd 1.620 1.098e-01
# or via confidence intervals
(LatM2 <- confint(paruelo.lmM2 <- lm(sqrt(C3) ~ cLAT *
c(cLONG + LongM2), data = paruelo))["cLAT", ]) 2.5 % 97.5 %
0.04830 0.09784 (LatM1 <- confint(paruelo.lmM2 <- lm(sqrt(C3) ~ cLAT *
c(cLONG + LongM1), data = paruelo))["cLAT", ]) 2.5 % 97.5 %
0.04220 0.07457 (LatM <- confint(paruelo.lmM2 <- lm(sqrt(C3) ~ cLAT *
c(cLONG + LongM), data = paruelo))["cLAT", ]) 2.5 % 97.5 %
0.03398 0.05340 (LatP1 <- confint(paruelo.lmM2 <- lm(sqrt(C3) ~ cLAT *
c(cLONG + LongP1), data = paruelo))["cLAT", ]) 2.5 % 97.5 %
0.01849 0.03952 (LatP2 <- confint(paruelo.lmM2 <- lm(sqrt(C3) ~ cLAT *
c(cLONG + LongP2), data = paruelo))["cLAT", ]) 2.5 % 97.5 %
-0.003313 0.031946 rbind(`Lattitude at Long-2sd` = LatM2, `Lattitude at Long-1sd` = LatM1,
`Lattitude at mean Long` = LatM, `Lattitude at Long+1sd` = LatP1,
`Lattitude at Long+2sd` = LatP2) 2.5 % 97.5 %
Lattitude at Long-2sd 0.048297 0.09784
Lattitude at Long-1sd 0.042196 0.07457
Lattitude at mean Long 0.033984 0.05340
Lattitude at Long+1sd 0.018492 0.03952
Lattitude at Long+2sd -0.003313 0.03195