Tutorial 8.2b - Comparing two populations (Bayesian)
14 Jan 2013
This tutorial will focus on the use of Bayesian (MCMC sampling) estimation to explore differences between two populations. Bayesian estimation is supported within R via two main packages (MCMCpack and MCMCglmm). These packages provide relatively simple R-like interfaces (particularly MCMCpack) to Bayesian routines and are therefore reasonably good starting points for those transitioning between traditional and Bayesian approaches.
BUGS (Bayesian inference Using Gibbs Sampling) is an algorithm and supporting language (resembling R) dedicated to performing the Gibbs sampling implementation of Markov Chain Monte Carlo method. Dialects of the BUGS language are implemented within two main projects;
- WinBUGS/OpenBUGS - written in component pascal and therefore originally Windows only. A less platform specific version (OpenBUGS - Windows and Linux via some component pascal libraries) is now being developed as the WinBUGS successor. Unlike WinBUGS, OpenBUGS does not have a Graphical User Interface and was designed to be invoked from other applications (such as R). The major drawback of WinBUGS/OpenBUGS is that it is relatively slow.
- JAGS (Just Another Gibbs Sampler) - written in C++ and is therefore cross-platform and very fast. It can also be called from within R via various packages.
Whilst the above programs can be used stand-alone, they do offer the rich data pre-processing and graphical capabilities of R, and thus, they are best accessed from within R itself. As such there are multiple packages devoted to interfacing with the various BUGS implementations;
- R2WinBUGS - interfaces with WinBUGS/OpenBUGS
- rjags - interfaces with JAGS
- R2jags - interfaces with JAGS yet also provides some of the more advanced post-processing routines offered within R2WinBUGS. Actually, it is sort of a hybrid between R2WinBUGS, rjags and coda packages.
There is an additional package (coda) that provides many routines for model checking and exploring the posterior distributions.
The BUGS language and algorithm is very powerful and flexible. However, the cost of this power and flexibility is complexity and the need for a firm understanding of the model you wish to fit as well as the priors to be used. The BUGS algorithm requires the following;
- the model comprising;
- likelihood function relating the response to the predictors - this is where good knowledge of the conceptual model that you are seeking to fit is essential.
- definition of the priors
- chain properties such;
- number of chains
- length of chains (number of iterations)
- burn-in length (number of initial iterations to ignore)
- thinning rate (number of iterations to count on before storing a sample)
- initial estimates to start an mcmc chain. If there are multiple chains, these starting values can differ between chains
- a list of model parameters and derivatives to monitor (and return the posterior distributions of)
We will start by generating a random data set. Note, I am creating two versions of the predictor variable (a numeric version and a factorial version).
> set.seed(1) > n1 <- 60 #sample size from population 1 > n2 <- 40 #sample size from population 2 > mu1 <- 105 #population mean of population 1 > mu2 <- 77.5 #population mean of population 2 > sigma <- 2.75 #standard deviation of both populations (equally varied) > n <- n1 + n2 #total sample size > y1 <- rnorm(n1, mu1, sigma) #population 1 sample > y2 <- rnorm(n2, mu2, sigma) #population 2 sample > y <- c(y1, y2) > x <- factor(rep(c("A", "B"), c(n1, n2))) #categorical listing of the populations > # xn.1 <- rep(c(0,1),c(n1,n2)) #numerical version of the population category for effects > # parameterization. Should start at 0. > xn <- as.numeric(x) #numerical version of the population category for means parameterization. Should not start at 0. > data <- data.frame(y, x, xn) # dataset > head(data) #print out the first six rows of the data set
y x xn 1 103.3 A 1 2 105.5 A 1 3 102.7 A 1 4 109.4 A 1 5 105.9 A 1 6 102.7 A 1
Lets begin by performing the usual exploratory data analysis - in this case, a boxplot of the response for each level of the predictor.
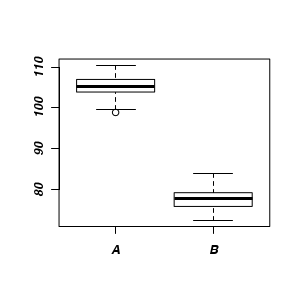 |
View code
> boxplot(y ~ x, data)
|
MCMCpack
> library(MCMCpack) > data.ttest <- MCMCregress(y ~ x, data = data)
Prior to inspecting any summaries of the parameter estimates, it is prudent to inspect a range of chain convergence diagnostics
- Trace plots
View trace plotsTrace plots show no evidence that the chains have not reasonably traversed the entire multidimensional parameter space.
> plot(data.ttest)
- Raftery diagnostic
View Raftery diagnosticThe Raftery diagnostics estimate that we would require about 3900 samples to reach the specified level of confidence in convergence. As we have 10,000 samples, we can be confidence that convergence has occurred.
> raftery.diag(data.ttest)Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 Burn-in Total Lower bound Dependence (M) (N) (Nmin) factor (I) (Intercept) 2 3802 3746 1.010 xB 2 3650 3746 0.974 sigma2 2 3865 3746 1.030 - Autocorrelation diagnostic
View autocorrelationsA lag of 1 appears to be mainly sufficient to avoid autocorrelation (except for sigma2, which is over 0.1). The diagnostic suggests that a lag of 5 would potentially correct this.
> autocorr.diag(data.ttest)(Intercept) xB sigma2 Lag 0 1.0000000 1.000000 1.000000 Lag 1 0.0095872 -0.001081 0.027941 Lag 5 0.0062995 0.011489 0.005293 Lag 10 0.0003455 0.011819 -0.002390 Lag 50 -0.0150021 -0.023501 -0.005571
Notwithstanding the slight issue of autocorrelation in the sigma2 samples, there is no evidence that the mcmc chain did not converge on a stable posterior distribution. We are now in a position to examine the summaries of the parameters.
> summary(data.ttest)
Iterations = 1001:11000
Thinning interval = 1
Number of chains = 1
Sample size per chain = 10000
1. Empirical mean and standard deviation for each variable,
plus standard error of the mean:
Mean SD Naive SE Time-series SE
(Intercept) 105.3 0.324 0.00324 0.00324
xB -27.5 0.516 0.00516 0.00516
sigma2 6.3 0.926 0.00926 0.00952
2. Quantiles for each variable:
2.5% 25% 50% 75% 97.5%
(Intercept) 104.67 105.08 105.30 105.51 105.93
xB -28.51 -27.83 -27.49 -27.15 -26.47
sigma2 4.75 5.65 6.21 6.86 8.33
The Group A is typically 27.5 units greater than Group B. The 95% confidence interval for the difference between Group A and B does not overlap with 0 implying a significant difference between the two groups. Compare this to a frequentist students' t-test.
We can also get the means and quantiles for each group. We will also add a p-value that corresponds to an empirical p-value for the hypothesis that the specified columns of samp have mean zero versus a general multivariate distribution with elliptical contours.
> # get the means of each group > tab <- rbind(Group1 = c(mean = mean(data.ttest[, 1]), quantile(data.ttest[, 1], c(0.025, 0.25, 0.5, 0.75, + 0.975))), Group2 = c(mean = mean(data.ttest[, 1] + data.ttest[, 2]), quantile(data.ttest[, 1] + data.ttest[, + 2], c(0.025, 0.25, 0.5, 0.75, 0.975)))) > tab
mean 2.5% 25% 50% 75% 97.5% Group1 105.3 104.67 105.08 105.30 105.51 105.93 Group2 77.8 77.03 77.54 77.81 78.07 78.58
Whilst workers attempt to become comfortable with a new statistical framework, it is only natural that they like to evaluate and comprehend new structures and output alongside more familiar concepts. One way to facilitate this is via Bayesian p-values that are somewhat analogous to the frequentist p-values for investigating the hypothesis that the two populations are identical (t=0).
> library(coda) > mcmcpvalue <- function(samp) { + ## elementary version that creates an empirical p-value for the hypothesis that the columns of samp + ## have mean zero versus a general multivariate distribution with elliptical contours. + + ## differences from the mean standardized by the observed variance-covariance factor + + ## Note, I put in the bit for single terms + if (length(dim(samp)) == 0) { + std <- backsolve(chol(var(samp)), cbind(0, t(samp)) - mean(samp), transpose = TRUE) + sqdist <- colSums(std * std) + sum(sqdist[-1] > sqdist[1])/length(samp) + } else { + std <- backsolve(chol(var(samp)), cbind(0, t(samp)) - colMeans(samp), transpose = TRUE) + sqdist <- colSums(std * std) + sum(sqdist[-1] > sqdist[1])/nrow(samp) + } + + } > mcmcpvalue(data.ttest[, 2])
[1] 0
With a p-value of 0, we would reject the frequentist null hypothesis of no difference between two population means.
JAGS
As indicated earlier, BUGS and therefore JAGS, requires more careful thought and effort to fit a model. In particular, we need to be very clear on the model being fitted so that we can correctly define the appropriate and corresponding likelihood function.
Broadly, there are two ways of parameterizing (expressing the unknown (to be estimated) components of a model) a model. Either we can estimate the means of each group (Means parameterization) or we can estimate the mean of one group and the difference between this group and the other group(s) (Effects parameterization). The latter is commonly used for frequentist null hypothesis testing as its parameters are more consistent with the null hypothesis of interest (that the difference between the two groups equals zero).
| Effects parameterization | Means parameterization |
|---|---|
|
yi = α + βj(i)*xi + εi where ε ∼ N(0,σ²)
|
yi = αj(i) + εi where ε ∼ N(0,σ²)
|
|
Each of the (i) values of y are modelled by an intercept (α - mean of Group A)
plus a difference parameter (β - difference between mean of Group A and Group B) multiplied by an indicator of which group the observation came from
plus a residual drawn from a normal distribution (N) with a mean of zero and a variance of σ². Actually, there are as many β parameters as there are groups (j), however one of them (typically the first) is set to be equal to zero (to avoid over-parameterization). |
Each of the (i) values of y are modelled as the mean (α) of each group (j) plus a residual drawn from a normal distribution (N) with a mean of zero and a variance of σ². Actually, α is a set of j coefficients corresponding to the j dummy coded factor levels. |
Alternatively, we can express these as:
| Effects parameterization | Means parameterization |
|---|---|
|
yi ~ N(α + βj(i)*xi, σ²)
|
yi ~ N(αj(i), σ²)
|
|
Each of the (i) values of y are modelled assuming they are drawn from a normal
distribution whose mean is determined by a linear combination of effect parameters
(α + βj(i)*xi)
and whose
variance is defined by the degree of variability (σ²) in this mean. There are three parameters;
|
Each of the (i) values of y are modelled assuming they are drawn from a normal
distribution whose mean is determined by a linear combination of means parameters
(&alphaj(i))
and whose
variance is defined by the degree of variability (σ²) in this mean.
There are three parameters;
|
Actually in the various BUGS dialects, distributions are defined by their precision (τ) rather than their variance (σ²). Precision is just the inverse of variance (τ = 1/σ²). Precision is used instead of variance as it permits the gamma distribution to be used as the conjugate prior of the variance of a normal distribution.
Bayesian analyses require that priors be specified for each of the parameters. We will define vague (non-informative) priors for each of the parameters such that the posterior distributions are almost entirely influenced by the likelihood (and thus the data). Hence, appropriate (conjugate) priors for the effects parameterization could be:
- α - a very flat (in-precise) normal distribution centered around zero, N(0, 1.0E-6). Note, 1.0E-6 is scientific notation for 0.000001
- β2 - a very flat (in-precise) normal distribution centered around zero, N(0,1.0E-6)
- τ - a very in-precise gamma distribution with a shape parameter close to zero (must be greater than 0)

Hence, the above equation becomes:
| Effects parameterization | Means parameterization |
|---|---|
|
yi ~ N(mui, τ)
mui = α + βj*xi τ = 1/σ² α ~ N(0, 1.0E-6) β1 = 0 β2 ~ N(0, 1.0E-6) σ ~ gamma(0.001, 0.001) |
yi ~ N(αj(i), σ²)
mui = α + βj*xi τ = 1/σ² α1 ~ N(0, 1.0E-6) α2 ~ N(0, 1.0E-6) σ ~ gamma(0.001, 0.001) |
|
The first two lines define the model likelihood. The third line defines the τ parameter in terms of variance. The next four lines define the priors for each parameter |
The first two lines define the model likelihood. The third line defines the τ parameter in terms of variance. The next three lines define the priors for each parameter. |
The BUGS language very closely matches the above model and prior definitions - hence the importance on understanding the model you wish to fit. The BUGS language resembles R in many respects. It basically consists of:
- stochastic nodes - those that appear on the left hand side of ~
- deterministic nodes - those that appear on the left hand side of <-
- R-like for loops
- R-like functions to transform and summarize data
- the order with which statements appear in the model definition are not important
- nodes should not be defined more than once (you cannot change a value
- We are now in a good position to define the model (Likelihood function and prior distributions).
Effects parameterization Means parameterization > modelString=" + model { + #Likelihood + for (i in 1:n) { + y[i]~dnorm(mu[i],tau) + mu[i] <- alpha+beta[x[i]] + } + + #Priors + alpha ~ dnorm (0,0.001) + beta[1] <- 0 + beta[2] ~ dnorm(0,0.001) + tau <- 1 / (sigma * sigma) + sigma~dgamma(0.001,0.001) + + #Other Derived parameters + # Group means (note, beta is a vector) + Group.means <-alpha+beta + } + "> modelString.means=" + model { + #Likelihood + for (i in 1:n) { + y[i]~dnorm(mu[i],tau) + mu[i] <- alpha[x[i]] + } + + #Priors + for (j in min(x):max(x)) { + alpha[j] ~ dnorm(0,0.001) + } + + tau <- 1 / (sigma * sigma) + sigma~dgamma(0.001,0.001) + + #Other Derived parameters + effect <-alpha[2]-alpha[1] + } + "- Effects
-
> ## write the model to a text file > writeLines(modelString, con = "../downloads/BUGSscripts/ttestModel.txt")
- Means
-
> ## write the model to a text file > writeLines(modelString.means, con = "../downloads/BUGSscripts/ttestModelMeans.txt")
-
Arrange the data as a list (as required by BUGS). Note, all variables must be numeric, therefore we use the numeric version of x.
Furthermore, the first level must be 1.- Effects
-
> data.list <- with(data, list(y = y, x = xn, n = nrow(data)))
- Means
-
> data.list.means <- with(data, list(y = y, x = xn, n = nrow(data)))
-
Define the initial values (α, β and σ2 for the chain.
Reasonable starting points can be gleaned from the data themselves.
- Effects
-
> inits <- list(alpha = mean(data$y), beta = c(0, diff(tapply(data$y, data$x, mean))), sigma = sd(data$y/2))
- Means
-
> inits.means <- list(alpha = tapply(data$y, data$x, mean), sigma = sd(data$y/2))
- Define the nodes (parameters and derivatives) to monitor
- Effects
-
> params <- c("alpha", "beta", "sigma", "Group.means")
- Means
-
> params.means <- c("alpha", "effect", "sigma")
-
Define the chain parameters
> adaptSteps = 1000 > burnInSteps = 2000 > nChains = 3 > numSavedSteps = 50000 > thinSteps = 1 > nIter = ceiling((numSavedSteps * thinSteps)/nChains) - Start the JAGS model (Check the model, load data into the model, specify the number of chains and compile the model.
- Effects
-
> library(rjags)> jagsModel <- jags.model("../downloads/BUGSscripts/ttestModel.txt", data = data.list, inits = inits, n.chains = nChains, + n.adapt = adaptSteps)
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 214 Initializing model
- Means
-
> library(rjags)> jagsModel.means <- jags.model("../downloads/BUGSscripts/ttestModelMeans.txt", data = data.list.means, + inits = inits.means, n.chains = nChains, n.adapt = adaptSteps)
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 211 Initializing model
-
Update the model
- Effects
-
> update(jagsModel, n.iter = burnInSteps) - Means
-
> update(jagsModel.means, n.iter = burnInSteps)
- Extract and summarize the samples
- Effects
-
> data.mcmc <- coda.samples(jagsModel, variable.names = params, n.iter = nIter, thin = thinSteps) > summary(data.mcmc)
Iterations = 3001:19667 Thinning interval = 1 Number of chains = 3 Sample size per chain = 16667 1. Empirical mean and standard deviation for each variable, plus standard error of the mean: Mean SD Naive SE Time-series SE Group.means[1] 105.3 0.324 0.001448 0.00222 Group.means[2] 77.8 0.396 0.001769 0.00176 alpha 105.3 0.324 0.001448 0.00222 beta[1] 0.0 0.000 0.000000 0.00000 beta[2] -27.5 0.512 0.002288 0.00348 sigma 2.5 0.182 0.000814 0.00107 2. Quantiles for each variable: 2.5% 25% 50% 75% 97.5% Group.means[1] 104.64 105.06 105.28 105.50 105.92 Group.means[2] 77.03 77.55 77.81 78.07 78.58 alpha 104.64 105.06 105.28 105.50 105.92 beta[1] 0.00 0.00 0.00 0.00 0.00 beta[2] -28.47 -27.81 -27.47 -27.13 -26.47 sigma 2.18 2.37 2.49 2.62 2.89 - Means
-
> data.mcmc.means <- coda.samples(jagsModel.means, variable.names = params.means, n.iter = nIter, thin = thinSteps) > summary(data.mcmc.means)
Iterations = 3001:19667 Thinning interval = 1 Number of chains = 3 Sample size per chain = 16667 1. Empirical mean and standard deviation for each variable, plus standard error of the mean: Mean SD Naive SE Time-series SE alpha[1] 105.3 0.322 0.001440 0.00144 alpha[2] 77.8 0.398 0.001780 0.00178 effect -27.5 0.514 0.002298 0.00230 sigma 2.5 0.181 0.000811 0.00109 2. Quantiles for each variable: 2.5% 25% 50% 75% 97.5% alpha[1] 104.65 105.07 105.29 105.50 105.92 alpha[2] 77.01 77.53 77.79 78.06 78.57 effect -28.50 -27.84 -27.49 -27.15 -26.48 sigma 2.18 2.37 2.49 2.62 2.89
MCMC diagnostics
Again, prior to examining the summaries, we should have explored the convergence diagnostics.
- Trace plots
Trace plots show no evidence that the chains have not reasonably traversed the entire multidimensional parameter space.
> plot(data.mcmc)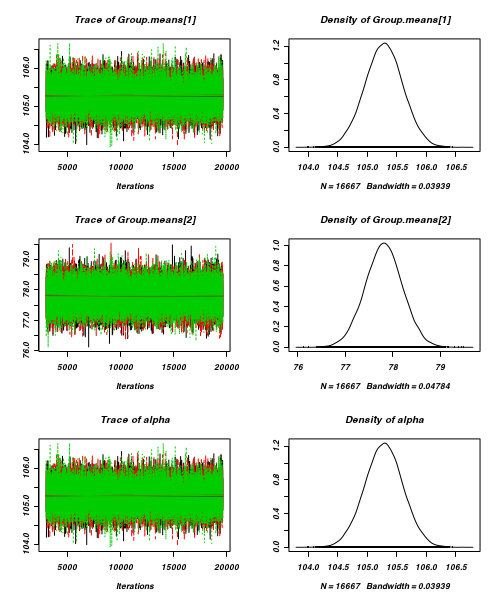
- Raftery diagnostic
The Raftery diagnostics for each chain estimate that we would require no more than 5000 samples to reach the specified level of confidence in convergence. As we have 16,667 samples, we can be confidence that convergence has occurred.
> raftery.diag(data.mcmc)[[1]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 Burn-in Total Lower bound Dependence (M) (N) (Nmin) factor (I) Group.means[1] 4 4676 3746 1.25 Group.means[2] 2 3912 3746 1.04 alpha 4 4676 3746 1.25 beta[1]3746 NA beta[2] 3 4560 3746 1.22 sigma 5 5563 3746 1.49 [[2]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 Burn-in Total Lower bound Dependence (M) (N) (Nmin) factor (I) Group.means[1] 3 4560 3746 1.22 Group.means[2] 2 3799 3746 1.01 alpha 3 4560 3746 1.22 beta[1] 3746 NA beta[2] 3 4493 3746 1.20 sigma 5 5607 3746 1.50 [[3]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 Burn-in Total Lower bound Dependence (M) (N) (Nmin) factor (I) Group.means[1] 4 4653 3746 1.24 Group.means[2] 2 3670 3746 0.98 alpha 4 4653 3746 1.24 beta[1] 3746 NA beta[2] 3 4538 3746 1.21 sigma 5 5694 3746 1.52 - Autocorrelation diagnostic
A lag of 1 appears insufficient to avoid autocorrelation (poor mixing). The diagnostic suggests that a thinning factor of 10 would potentially correct this.
> autocorr.diag(data.mcmc)Group.means[1] Group.means[2] alpha beta[1] beta[2] sigma Lag 0 1.000000 1.0000000 1.000000 NaN 1.000000 1.000000 Lag 1 0.402997 0.0056980 0.402997 NaN 0.399833 0.273830 Lag 5 0.005506 -0.0028340 0.005506 NaN 0.009176 -0.008363 Lag 10 -0.001560 0.0049169 -0.001560 NaN 0.005375 0.002866 Lag 50 -0.009238 -0.0005035 -0.009238 NaN -0.004914 0.003003
- Re-run the chain with a thinning factor of 10 and confirm better mixing
View codeThinning interval (lag) of 10, seems appropriate, although this has very little impact in this very simple case.
> jagsModel <- jags.model("../downloads/BUGSscripts/ttestModel.txt", data = data.list, inits = inits, n.chains = nChains, + n.adapt = adaptSteps)
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 214 Initializing model
> update(jagsModel, n.iter = burnInSteps) > data.mcmc <- coda.samples(jagsModel, variable.names = params, n.iter = nIter, thin = 10) > autocorr.diag(data.mcmc)
Group.means[1] Group.means[2] alpha beta[1] beta[2] sigma Lag 0 1.0000000 1.000000 1.0000000 NaN 1.0000000 1.000000 Lag 10 0.0068958 0.012882 0.0068958 NaN 0.0008962 0.009731 Lag 50 -0.0056822 -0.009699 -0.0056822 NaN -0.0183367 -0.012576 Lag 100 0.0008555 0.012004 0.0008555 NaN -0.0034160 0.007553 Lag 500 0.0033313 -0.013886 0.0033313 NaN -0.0215536 -0.027307
> summary(data.mcmc)Iterations = 3010:19670 Thinning interval = 10 Number of chains = 3 Sample size per chain = 1667 1. Empirical mean and standard deviation for each variable, plus standard error of the mean: Mean SD Naive SE Time-series SE Group.means[1] 105.3 0.324 0.00458 0.00448 Group.means[2] 77.8 0.392 0.00555 0.00557 alpha 105.3 0.324 0.00458 0.00448 beta[1] 0.0 0.000 0.00000 0.00000 beta[2] -27.5 0.511 0.00722 0.00722 sigma 2.5 0.184 0.00260 0.00260 2. Quantiles for each variable: 2.5% 25% 50% 75% 97.5% Group.means[1] 104.65 105.05 105.27 105.49 105.9 Group.means[2] 77.03 77.55 77.80 78.07 78.6 alpha 104.65 105.05 105.27 105.49 105.9 beta[1] 0.00 0.00 0.00 0.00 0.0 beta[2] -28.46 -27.82 -27.46 -27.12 -26.5 sigma 2.17 2.37 2.49 2.62 2.9
R2jags
As indicated earlier, we can alternatively access JAGS from functions within the R2jags package. Doing so provides some additional information. Notably we can obtain estimates of Deviance (and therefore DIC) as well as effective sample sizes for each of the parameters.
> library(R2jags)
When using the jags() function (R2jags package), it is not necessary to provide initial values. However, if they are to be supplied, the inital values list must be a list the same length as the number of chains.
> data.r2jags <- jags(data=data.list, + inits=NULL, #or inits=list(inits,inits,inits) # since there are three chains + parameters.to.save=params, + model.file="../downloads/BUGSscripts/ttestModelR2JAGS.txt", + n.chains=3, + n.iter=nIter, + n.burnin=burnInSteps, + n.thin=10 + ) > print(data.r2jags)
Notes:
- If inits=NULL the jags() function will generate vaguely sensible initial values for each chain based on the data
- In addition to the mean and quantiles of each of the sample nodes, the jags() function will calculate:
- the effective sample size for each sample - if n.eff for a node is substantially less than the number of iterations, then it suggests poor mixing
- Rhat values for each sample - these are a convergence diagnostic (values of 1 indicate full convergence, values greater than 1.01 are indicative of non-convergence.
- an information criteria (DIC) for model selection
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 214 Initializing model
Inference for Bugs model at "../downloads/BUGSscripts/ttestModelR2JAGS.txt", fit using jags,
3 chains, each with 16667 iterations (first 2000 discarded), n.thin = 10
n.sims = 4401 iterations saved
mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff
Group.means[1] 105.283 0.322 104.642 105.069 105.278 105.494 105.925 1.001 4400
Group.means[2] 77.802 0.397 77.030 77.533 77.800 78.069 78.584 1.001 4400
alpha 105.283 0.322 104.642 105.069 105.278 105.494 105.925 1.001 4400
beta[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1
beta[2] -27.481 0.507 -28.471 -27.811 -27.489 -27.145 -26.498 1.001 4400
sigma 2.502 0.184 2.174 2.374 2.493 2.619 2.894 1.001 4400
deviance 466.706 2.468 463.866 464.880 466.080 467.840 473.134 1.001 4400
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 3.0 and DIC = 469.8
DIC is an estimate of expected predictive error (lower deviance is better).
> modelString="
+ model {
+ #Likelihood
+ for (i in 1:n) {
+ y[i]~dnorm(mu[i],tau)
+ mu[i] <- alpha+beta[x[i]]
+ }
+
+ #Priors
+ alpha ~ dnorm (0,0.001)
+ beta[1] <- 0
+ beta[2] ~ dnorm(0,0.001)
+ tau <- 1 / (sigma * sigma)
+ sigma~dgamma(0.001,0.001)
+
+ #Other Derived parameters
+ # Group means (note, beta is a vector)
+ Group.means <-alpha+beta
+ }
+ "
> writeLines(modelString, con="../downloads/BUGSscripts/ttestModelR2JAGS.txt")
>
> data.r2jags <- jags(data=data.list,
+ inits=NULL, #or inits=list(inits,inits,inits) # since there are three chains
+ parameters.to.save=params,
+ model.file="../downloads/BUGSscripts/ttestModelR2JAGS.txt",
+ n.chains=3,
+ n.iter=nIter,
+ n.burnin=burnInSteps,
+ n.thin=10
+ )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 214 Initializing model
> print(data.r2jags)
Inference for Bugs model at "../downloads/BUGSscripts/ttestModelR2JAGS.txt", fit using jags,
3 chains, each with 16667 iterations (first 2000 discarded), n.thin = 10
n.sims = 4401 iterations saved
mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff
Group.means[1] 105.280 0.330 104.636 105.06 105.278 105.50 105.93 1.001 4400
Group.means[2] 77.816 0.398 77.035 77.55 77.819 78.08 78.61 1.001 4400
alpha 105.280 0.330 104.636 105.06 105.278 105.50 105.93 1.001 4400
beta[1] 0.000 0.000 0.000 0.00 0.000 0.00 0.00 1.000 1
beta[2] -27.464 0.518 -28.497 -27.81 -27.465 -27.11 -26.44 1.001 4400
sigma 2.506 0.182 2.186 2.38 2.494 2.62 2.90 1.001 3700
deviance 466.725 2.543 463.844 464.89 466.037 467.92 473.37 1.001 4400
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 3.2 and DIC = 470.0
DIC is an estimate of expected predictive error (lower deviance is better).
The total number samples collected is 4401. That is, there are 4401 samples collected from the multidimensional posterior distribution and thus, 4401 samples collected from the posterior distributions of each parameter. The effective number of samples column indicates the number of independent samples represented in the total. It is clear that for all except sigma, the chains were well mixed. The number of effective samples of sigma was 1200, potentially indicating incomplete mixing.
From now on, we will perform all Bayesian analyses via jags() so as to leverage the maximum amount of knowledge available from the models.
Effect sizes
In addition to deriving the distribution means for the second group, we could make use of the Bayesian framework to derive the distribution of the effect size. There are multiple ways of calculating an effect size, but the two most common are:
- Raw effect size
- the difference between two groups (as already calculated)
- Cohen's D
- the effect size standardized by division with the pooled standard deviation
= (μA - μB)/σ
- Percent effect size
- expressing the effect size as a percent of the reference group mean
> modelString="
+ model {
+ #Likelihood
+ for (i in 1:n) {
+ y[i]~dnorm(mu[i],tau)
+ mu[i] <- alpha+beta[x[i]]
+ }
+
+ #Priors
+ alpha ~ dnorm(0.001,1.0E-6)
+ beta[1] <- 0
+ beta[2] ~ dnorm(0,1.0E-6)
+
+ tau <- 1 / (sigma * sigma)
+ sigma~dgamma(0.001,0.001)
+
+ #Other Derived parameters
+ Group.means <-alpha+beta # Group means
+
+ cohenD <- beta[2]/sigma
+ ES <- 100*beta[2]/alpha
+ }
+ "
Notes:
- Calculating the percent effect size involves division by an estimate of α. The very first sample collected of each parameter (including α) is based on the initial values supplied.
If inits=NULL the jags() function appears to generate initial values from the priors.
Recall that in the previous model definition, α was deemed to be distributed as a normal distribution with a mean of 0.
Hence, α would initially be assigned a value of 0. Division by zero is of course illegal and thus an error would be thrown.
There are two ways to overcome this:
- modify the prior such that it has a mean of close to zero (and thus the first α sample is not zero), yet not actually zero (such as 0.0001). There is always a danger however that the next sample in the chain will be zero (although this is highly unlikely).
- define initial values that are based on the observed data (and not zero). This is the method used here - see the initial values used.
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 219 Initializing model
Inference for Bugs model at "../downloads/BUGSscripts/R2JAGSEffectSize.txt", fit using jags,
3 chains, each with 16667 iterations (first 2000 discarded), n.thin = 10
n.sims = 4401 iterations saved
mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff
ES -26.111 0.441 -26.974 -26.407 -26.104 -25.817 -25.251 1.001 4400
Group.means[1] 105.300 0.325 104.654 105.086 105.294 105.525 105.936 1.001 4400
Group.means[2] 77.804 0.399 77.035 77.533 77.807 78.077 78.558 1.001 4400
alpha 105.300 0.325 104.654 105.086 105.294 105.525 105.936 1.001 4400
beta[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1
beta[2] -27.496 0.513 -28.507 -27.841 -27.484 -27.158 -26.514 1.001 4400
cohenD -11.050 0.808 -12.711 -11.595 -11.041 -10.496 -9.481 1.001 2600
sigma 2.501 0.178 2.182 2.375 2.489 2.616 2.873 1.001 2800
deviance 466.676 2.469 463.862 464.900 466.061 467.789 472.954 1.001 4400
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 3.0 and DIC = 469.7
DIC is an estimate of expected predictive error (lower deviance is better).
> modelString="
+ model {
+ #Likelihood
+ for (i in 1:n) {
+ y[i]~dnorm(mu[i],tau)
+ mu[i] <- alpha+beta[x[i]]
+ }
+
+ #Priors
+ alpha ~ dnorm(0.001,1.0E-6)
+ beta[1] <- 0
+ beta[2] ~ dnorm(0,1.0E-6)
+
+ tau <- 1 / (sigma * sigma)
+ sigma~dgamma(0.001,0.001)
+
+ #Other Derived parameters
+ Group.means <-alpha+beta # Group means
+
+ cohenD <- beta[2]/sigma
+ ES <- 100*beta[2]/alpha
+ }
+ "
> writeLines(modelString,con="../downloads/BUGSscripts/R2JAGSEffectSize.txt")
>
> data.list <- with(data,
+ list(y=y,
+ x=xn,n=nrow(data))
+ )
> inits <- list(alpha=mean(data$y),
+ beta=c(0,diff(tapply(data$y,data$x,mean))),
+ sigma=sd(data$y/2))
> params <- c("alpha","beta","sigma","Group.means","cohenD","ES")
> adaptSteps = 1000
> burnInSteps = 2000
> nChains = 3
> numSavedSteps = 50000
> thinSteps = 1
> nIter = ceiling((numSavedSteps * thinSteps)/nChains)
>
> data.r2jagsES <- jags(data=data.list,
+ inits=list(inits,inits,inits), # since there are three chains
+ parameters.to.save=params,
+ model.file="../downloads/BUGSscripts/R2JAGSEffectSize.txt",
+ n.chains=3,
+ n.iter=nIter,
+ n.burnin=burnInSteps,
+ n.thin=10
+ )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 219 Initializing model
> print(data.r2jagsES)
Inference for Bugs model at "../downloads/BUGSscripts/R2JAGSEffectSize.txt", fit using jags,
3 chains, each with 16667 iterations (first 2000 discarded), n.thin = 10
n.sims = 4401 iterations saved
mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff
ES -26.116 0.441 -26.967 -26.418 -26.11 -25.823 -25.249 1.001 4400
Group.means[1] 105.300 0.322 104.676 105.081 105.30 105.515 105.939 1.001 4200
Group.means[2] 77.799 0.399 77.012 77.527 77.80 78.066 78.584 1.001 4400
alpha 105.300 0.322 104.676 105.081 105.30 105.515 105.939 1.001 4200
beta[1] 0.000 0.000 0.000 0.000 0.00 0.000 0.000 1.000 1
beta[2] -27.501 0.513 -28.496 -27.851 -27.49 -27.161 -26.490 1.001 4400
cohenD -11.051 0.806 -12.678 -11.584 -11.05 -10.513 -9.504 1.002 1600
sigma 2.501 0.178 2.174 2.382 2.49 2.609 2.886 1.002 1400
deviance 466.651 2.453 463.851 464.861 465.98 467.764 473.083 1.001 4200
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 3.0 and DIC = 469.7
DIC is an estimate of expected predictive error (lower deviance is better).
The Cohen's D value is -11.05.
This value is far greater than the nominal 'large effect' guidelines outlined by Cohen
and thus we might proclaim the treatment as having a large negative effect.
The effect size expressed as a percentage of the Group A mean is -26.12.
Hence the treatment was associated with a 26.1% reduction.
Probability statements
Bayesian statistics provide a natural means to generate probability statements. For example, we could calculate the probability that there is an effect of the treatment. Moreover, we could calculate the probability that the treatment effect exceeds some threshold (which might be based on a measure of ecologically important difference or other compliance guidelines for example).
> modelString="
+ model {
+ #Likelihood
+ for (i in 1:n) {
+ y[i]~dnorm(mu[i],tau)
+ mu[i] <- alpha+beta[x[i]]
+ }
+
+ #Priors
+ alpha ~ dnorm(0.001,1.0E-6)
+ beta[1] <- 0
+ beta[2] ~ dnorm(0,1.0E-6)
+
+ tau <- 1 / (sigma * sigma)
+ sigma~dgamma(0.001,0.001)
+
+ #Other Derived parameters
+ Group.means <-alpha+beta # Group means
+
+ cohenD <- beta[2]/sigma
+ ES <- 100*beta[2]/alpha
+ P0 <- 1-step(beta[2])
+ P25 <- 1-step(ES+25)
+ }
+ "
Notes:
- We have defined two additional probability derivatives, both of which utilize the step function (which generates a binary vector based on whether values evaluate less than zero or not):
- P0 - the probability (mean of 1-step()) that the raw effect is greater than zero.
- P25 - the probability (mean of 1-step()) that the percent effect size is greater than 25%.
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 225 Initializing model
Inference for Bugs model at "../downloads/BUGSscripts/R2JAGSProbability.txt", fit using jags,
3 chains, each with 16667 iterations (first 2000 discarded), n.thin = 10
n.sims = 4401 iterations saved
mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff
ES -26.101 0.440 -26.973 -26.392 -26.101 -25.805 -25.226 1.001 4400
Group.means[1] 105.292 0.325 104.638 105.072 105.293 105.508 105.933 1.001 4400
Group.means[2] 77.809 0.398 77.036 77.546 77.807 78.071 78.574 1.001 4400
P0 1.000 0.000 1.000 1.000 1.000 1.000 1.000 1.000 1
P25 0.995 0.074 1.000 1.000 1.000 1.000 1.000 1.046 1800
alpha 105.292 0.325 104.638 105.072 105.293 105.508 105.933 1.001 4400
beta[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1
beta[2] -27.483 0.511 -28.491 -27.820 -27.476 -27.147 -26.483 1.001 4400
cohenD -11.039 0.811 -12.639 -11.582 -11.039 -10.494 -9.466 1.001 4400
sigma 2.502 0.180 2.179 2.377 2.489 2.613 2.896 1.001 4400
deviance 466.673 2.507 463.855 464.826 466.025 467.834 473.167 1.001 2700
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 3.1 and DIC = 469.8
DIC is an estimate of expected predictive error (lower deviance is better).
> modelString="
+ model {
+ #Likelihood
+ for (i in 1:n) {
+ y[i]~dnorm(mu[i],tau)
+ mu[i] <- alpha+beta[x[i]]
+ }
+
+ #Priors
+ alpha ~ dnorm(0.001,1.0E-6)
+ beta[1] <- 0
+ beta[2] ~ dnorm(0,1.0E-6)
+
+ tau <- 1 / (sigma * sigma)
+ sigma~dgamma(0.001,0.001)
+
+ #Other Derived parameters
+ Group.means <-alpha+beta # Group means
+
+ cohenD <- beta[2]/sigma
+ ES <- 100*beta[2]/alpha
+ P0 <- 1-step(beta[2])
+ P25 <- 1-step(ES+25)
+ }
+ "
> writeLines(modelString,con="../downloads/BUGSscripts/R2JAGSProbability.txt")
>
> data.list <- with(data,
+ list(y=y,
+ x=xn,n=nrow(data))
+ )
> inits <- list(alpha=mean(data$y),
+ beta=c(0,diff(tapply(data$y,data$x,mean))),
+ sigma=sd(data$y/2))
> params <- c("alpha","beta","sigma","Group.means","cohenD","ES","P0","P25")
> adaptSteps = 1000
> burnInSteps = 2000
> nChains = 3
> numSavedSteps = 5000
> thinSteps = 10
> nIter = ceiling((numSavedSteps * thinSteps)/nChains)
>
> data.r2jagsProb <- jags(data=data.list,
+ inits=list(inits,inits,inits), # since there are three chains
+ parameters.to.save=params,
+ model.file="../downloads/BUGSscripts/R2JAGSProbability.txt",
+ n.chains=3,
+ n.iter=nIter,
+ n.burnin=burnInSteps,
+ n.thin=10
+ )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 225 Initializing model
> print(data.r2jagsProb)
Inference for Bugs model at "../downloads/BUGSscripts/R2JAGSProbability.txt", fit using jags,
3 chains, each with 16667 iterations (first 2000 discarded), n.thin = 10
n.sims = 4401 iterations saved
mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff
ES -26.101 0.445 -26.974 -26.408 -26.101 -25.793 -25.223 1.001 4400
Group.means[1] 105.284 0.316 104.661 105.071 105.281 105.494 105.898 1.001 2400
Group.means[2] 77.803 0.407 77.001 77.529 77.809 78.073 78.591 1.001 4400
P0 1.000 0.000 1.000 1.000 1.000 1.000 1.000 1.000 1
P25 0.993 0.085 1.000 1.000 1.000 1.000 1.000 1.009 4400
alpha 105.284 0.316 104.661 105.071 105.281 105.494 105.898 1.001 2400
beta[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1
beta[2] -27.481 0.514 -28.484 -27.833 -27.476 -27.128 -26.473 1.001 4400
cohenD -11.059 0.817 -12.682 -11.607 -11.041 -10.482 -9.551 1.001 4400
sigma 2.498 0.180 2.179 2.372 2.489 2.614 2.863 1.001 4400
deviance 466.673 2.429 463.850 464.905 466.075 467.786 472.821 1.001 4400
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 3.0 and DIC = 469.6
DIC is an estimate of expected predictive error (lower deviance is better).
Finite population standard deviations
Finite-population standard deviationsIt is often useful to be able to estimate the relative amount of variability associated with each predictor (or term) in a model. This can provide a sort of relative importance measure for each predictor.
In frequentist statistics, such measures are only available for so called random factors (factors whose observational levels are randomly selected to represent all possible levels rather than to represent specific treatment levels). For such random factors, the collective variances (or standard deviation) of each factor are known as the variance components. Each component can also be expressed as a percentage of the total so as to provide a percentage breakdown of the relative contributions of each scale of sampling.
Frequentist approaches model random factors according to the variance they add to the model, whereas fixed factors are modelled according to their effects (deviations from reference means). The model does not seek to generalize beyond the observed levels of a given fixed factor (such as control vs treatment) and thus it apparently does not make sense to estimate the population variability between levels (which is what variance components estimate).
The notion of 'fixed' and 'random' factors is somewhat arbitrary and does not really have any meaning within a Bayesian context (as all parameters and thus factors are considered random). Instead, the spirit of what many consider is the difference between fixed and random factors can be captured by conceptualizing whether the levels of a factor are drawn from a finite-population (from which the observed factor levels are the only ones possible) or a superpopulation (from which the observed factor levels are just a random selection of the infinite possible levels possible).
Hence, variance components could be defining in terms of either finite-population or superpopulation standard deviations. Superpopulation standard deviations have traditionally been used to describe the relative scale of sampling variation (e.g. where is the greatest source of variability; Plots, subplots within plots, individual quadrats within subplots, .... or years, months within years, weeks within months, days within weeks, ...) and are most logically applicable to factors that have a relatively large number of levels (such as spatial or temporal sampling units). On the other hand, finite-population standard deviations can be used to explore the relative impact or effect of a set of (fixed) treatments.
- Calculate the amount of unexplained (residual) variance absorbed by the factor.
This is generated by fitting a model with (full model) and without (reduced model)
the term and subtracting the
standard deviations of the residuals one another.
σA = σReducedResid - σFullResidThis approach works fine for models that only include fixed factors (indeed it is somewhat analogous to the partitioning of variance employed by an ANOVA table), but cannot be used when the model includes random factors.View code if you are interested
> data.lmFull <- lm(y ~ x, data) > data.lmRed <- lm(y ~ 1, data) > sd.a <- sd(data.lmRed$resid) - sd(data.lmFull$resid) > sd.resid <- sd(data.lmFull$resid) > sds <- c(sd.a, sd.resid) > 100 * sds/sum(sds)
[1] 82.05 17.95
The Bayesian framework provides a relatively simple way to generate both finite-population and superpopulation standard deviation estimates for all factors.
- finite-population
- the standard deviations of the Markov Chain Monte Carlo samples across each of the parameters associated with a factor (eg, β1 and β2 in the effects parameterization model) provide natural estimates of the variability between group levels (and thus the finite-population standard deviation).
- superpopulation
-
the mechanism of defining priors also provides a mechanism for calculating
infinite-population standard deviations. Recall that in the cell means model,
the prior for α specifies that each of the α values are drawn from a normal distribution
with a particular mean and a certain level of precision (reciprocal of variability).
We could further parameterize this prior into an estimatable mean and precision via hyperpriors
αi ~ N(μ, τ)Since the normal distribution in line one above represents the distribution from which the (infinite) population means are drawn, τ (1/σ²) provides a direct measure of the variability of the population from which the means are drawn.
μ ~ N(0, 1.0E-6)
τ ~ gamma(0.001, 0.001)
When the number of levels of a factor a large, the finite-population and superpopulation standard deviation point estimates will be very similar. However, the finite-population estimates will of course be considered more precise (and thus, less varied). At the other extreme, when the number of factor levels is small (such as two levels), the finite-population estimate will be very precise whereas the superpopulation standard deviation estimate will be very imprecise (highly varied). For this reason, if the purpose of estimating standard deviations is to compare relative contributions of various predictors some of which have small numbers of levels and others large), then it is best to use finite-population standard deviation estimates.
> modelString="
+ model {
+ #Likelihood
+ for (i in 1:n) {
+ y[i]~dnorm(mu[i],tau)
+ mu[i] <- alpha+beta[x[i]]
+ resid[i] <- y[i]-mu[i]
+ }
+
+ #Priors
+ alpha ~ dnorm(0,1.0E-6)
+ beta[1] <- 0
+ beta[2] ~ dnorm(0,tau.a)
+
+ tau <- 1 / (sigma * sigma)
+ sigma~dgamma(0.001,0.001)
+
+ tau.a <- 1 / (sigma.a * sigma.a)
+ sigma.a~dgamma(0.001,0.001)
+
+ #Finite-population standard deviations
+ sd.a <- sd(beta)
+ sd.resid <- sd(resid)
+ }
+ "
Notes:
- The fixed precision constant of 1.0E-6 in the prior for β2 has been replaced by a parameter representing the modelled amount of precision (and thus standard deviation) of the normal distribution.
- Finite-population standard deviation derivatives are included for the treatment (sd.a) and the residuals (sd.resid).
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 320 Initializing model
Inference for Bugs model at "../downloads/BUGSscripts/R2JAGSSD.txt", fit using jags,
3 chains, each with 16667 iterations (first 2000 discarded), n.thin = 10
n.sims = 4401 iterations saved
mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff
alpha 105.302 0.326 104.665 105.078 105.310 105.522 105.931 1.001 4400
beta[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1
beta[2] -27.498 0.518 -28.530 -27.837 -27.498 -27.160 -26.483 1.001 4400
sd.a 13.749 0.259 13.241 13.580 13.749 13.919 14.265 1.001 4400
sd.resid 2.471 0.018 2.458 2.459 2.463 2.475 2.522 1.002 3100
sigma 2.501 0.177 2.180 2.375 2.492 2.619 2.869 1.003 890
sigma.a 73.253 107.699 12.025 23.455 38.448 74.680 391.479 1.001 2500
deviance 466.623 2.434 463.845 464.826 466.016 467.739 472.916 1.001 4400
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 3.0 and DIC = 469.6
DIC is an estimate of expected predictive error (lower deviance is better).
> modelString="
+ model {
+ #Likelihood
+ for (i in 1:n) {
+ y[i]~dnorm(mu[i],tau)
+ mu[i] <- alpha+beta[x[i]]
+ resid[i] <- y[i]-mu[i]
+ }
+
+ #Priors
+ alpha ~ dnorm(0,1.0E-6)
+ beta[1] <- 0
+ beta[2] ~ dnorm(0,tau.a)
+
+ tau <- 1 / (sigma * sigma)
+ sigma~dgamma(0.001,0.001)
+
+ tau.a <- 1 / (sigma.a * sigma.a)
+ sigma.a~dgamma(0.001,0.001)
+
+ #Finite-population standard deviations
+ sd.a <- sd(beta)
+ sd.resid <- sd(resid)
+ }
+ "
> writeLines(modelString,con="../downloads/BUGSscripts/R2JAGSSD.txt")
>
> data.list <- with(data,
+ list(y=y,
+ x=xn,n=nrow(data))
+ )
> inits <- list(alpha=mean(data$y),
+ beta=c(0,diff(tapply(data$y,data$x,mean))),
+ sigma=sd(data$y/2))
> params <- c("alpha","beta","sigma","sd.a","sd.resid","sigma.a")
> adaptSteps = 1000
> burnInSteps = 2000
> nChains = 3
> numSavedSteps = 50000
> thinSteps = 1
> nIter = ceiling((numSavedSteps * thinSteps)/nChains)
>
> data.r2jagsSD <- jags(data=data.list,
+ inits=list(inits,inits,inits), # since there are three chains
+ parameters.to.save=params,
+ model.file="../downloads/BUGSscripts/R2JAGSSD.txt",
+ n.chains=3,
+ n.iter=nIter,
+ n.burnin=burnInSteps,
+ n.thin=10
+ )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 320 Initializing model
> print(data.r2jagsSD)
Inference for Bugs model at "../downloads/BUGSscripts/R2JAGSSD.txt", fit using jags,
3 chains, each with 16667 iterations (first 2000 discarded), n.thin = 10
n.sims = 4401 iterations saved
mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff
alpha 105.29 0.320 104.653 105.077 105.301 105.512 105.919 1.001 4400
beta[1] 0.00 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1
beta[2] -27.48 0.507 -28.463 -27.821 -27.491 -27.139 -26.470 1.003 1100
sd.a 13.74 0.254 13.235 13.569 13.746 13.910 14.231 1.003 1100
sd.resid 2.47 0.017 2.458 2.459 2.463 2.475 2.521 1.001 4400
sigma 2.50 0.182 2.179 2.371 2.490 2.614 2.894 1.001 4400
sigma.a 71.14 120.908 11.787 22.860 37.871 70.985 354.121 1.002 4400
deviance 466.64 2.408 463.810 464.847 466.053 467.787 472.816 1.002 2400
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 2.9 and DIC = 469.5
DIC is an estimate of expected predictive error (lower deviance is better).
The between group (finite population) standard deviation is
13.74 whereas the within group
standard deviation is 2.47.
These equate to 84.8%, 15.2% respectively.
Compared to the finite-population standard deviation, the superpopulation between group standard deviation
estimate (σa) is both very large and highly variable (37.87, 2.49).
This is to be expected, whilst the finite-population standard deviation represents the degree
of variation between the observed levels, the superpopulation standard deviation seeks to
estimate the variability of the population from which the group means of the
observed levels AND all other possible levels are drawn. There are only two levels from which
to estimate this standard deviation and therefore, its value and variability are going to be
higher than those pertaining only to the scope of the current data.
Examination of the quantiles for σa suggest that its samples are not
distributed normally. Consequently, the mean is not an appropriate measure of its location.
We will instead characterize the superpopulation between group and within group standard deviations
via their respective medians (37.87, 2.49) and as percent medians (93.8%, 6.2% ).
The contrast between finite-population and superpopulation standard deviations is also emphasized by the respective estimates for the residuals. The residuals are of course a 'random' factor with a large number of observed levels. It is therefore not surprising that the point estimates for the residuals variance components are very similar. However, also notice that the precision of the finite-population standard deviation estimate is substantially higher (lower standard deviation of the standard deviation estimate) than that of the superpopulation estimate.
Unequally varied populations
> set.seed(1) > n1 <- 60 #sample size from population 1 > n2 <- 40 #sample size from population 2 > mu1 <- 105 #population mean of population 1 > mu2 <- 77.5 #population mean of population 2 > sigma1 <- 3 #standard deviation of population 1 > sigma2 <- 2 #standard deviation of population 2 > n <- n1 + n2 #total sample size > y1 <- rnorm(n1, mu1, sigma1) #population 1 sample > y2 <- rnorm(n2, mu2, sigma2) #population 2 sample > y <- c(y1, y2) > x <- factor(rep(c("A", "B"), c(n1, n2))) #categorical listing of the populations > xn <- rep(c(0, 1), c(n1, n2)) #numerical version of the population category > data2 <- data.frame(y, x, xn) # dataset > head(data2) #print out the first six rows of the data set
y x xn 1 103.1 A 0 2 105.6 A 0 3 102.5 A 0 4 109.8 A 0 5 106.0 A 0 6 102.5 A 0
- Start by defining the model
yi = α + β*xi + εiwhich in BUGS becomes;yi = α + β*xi + εi
ε1 ∼ N(0,τ1) for x1=0 (females)
ε2 ∼ N(0,τ2) for x2=1 (males)> modelString=" + model { + #Likelihood + for (i in 1:n1) { + y1[i]~dnorm(mu1,tau1) + } + for (i in 1:n2) { + y2[i]~dnorm(mu2,tau2) + } + + #Priors + mu1 ~ dnorm (0,0.001) + mu2 ~ dnorm(0,0.001) + tau1 <- 1 / (sigma1 * sigma1) + sigma1~dgamma(0.001,0.001) + tau2 <- 1 / (sigma2 * sigma2) + sigma2~dgamma(0.001,0.001) + + #Other Derived parameters + delta <- mu2 - mu1 + } + " > ## write the model to a text file (I suggest you alter the path to somewhere more relevant > ## to your system!) > writeLines(modelString,con="../downloads/BUGSscripts/ttestModel2.txt") > > > > modelString=" + model { + #Likelihood + for (i in 1:n1) { + y1[i]~dnorm(mu1,tau1) + } + for (i in 1:n2) { + y2[i]~dnorm(mu2,tau2) + } + + #Priors + mu1 ~ dnorm (0,0.001) + mu2 ~ dnorm(0,0.001) + tau1 <- 1 / (sigma1 * sigma1) + sigma1~dgamma(0.001,0.001) + tau2 <- 1 / (sigma2 * sigma2) + sigma2~dgamma(0.001,0.001) + + #Other Derived parameters + delta <- mu2 - mu1 + } + " > ## write the model to a text file (I suggest you alter the path to somewhere more relevant > ## to your system!) > writeLines(modelString,con="../downloads/BUGSscripts/ttestModel2.txt") -
Arrange the data as a list (as required by BUGS). Note, all variables must be numeric, therefore we use the numeric version of x.
Define the initial values. We are going to initialize three chains so the list must include three elements.
We will also define the other properties.
> data2.list <- with(data2, list(y1 = y[xn == 0], y2 = y[xn == 1], n1 = length(y[xn == 0]), n2 = length(y[xn == + 1]))) > inits <- list(list(mu1 = rnorm(1), mu2 = rnorm(1), sigma1 = rlnorm(1), sigma2 = rlnorm(1)), list(mu1 = rnorm(1), + mu2 = rnorm(1), sigma1 = rlnorm(1), sigma2 = rlnorm(1)), list(mu1 = rnorm(1), mu2 = rnorm(1), sigma1 = rlnorm(1), + sigma2 = rlnorm(1))) > params <- c("mu1", "mu2", "delta", "sigma1", "sigma2") > adaptSteps = 1000 > burnInSteps = 2000 > nChains = 3 > numSavedSteps = 50000 > thinSteps = 1 > nIter = ceiling((numSavedSteps * thinSteps)/nChains)
-
Engage jags
> library(R2jags) > data2.r2jags <- jags(data=data2.list, + inits=NULL, #or inits=list(inits,inits,inits) # since there are three chains + parameters.to.save=params, + model.file="../downloads/BUGSscripts/ttestModel2.txt", + n.chains=3, + n.iter=nIter, + n.burnin=burnInSteps, + n.thin=10 + )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 114 Initializing model
> print(data2.r2jags)Inference for Bugs model at "../downloads/BUGSscripts/ttestModel2.txt", fit using jags, 3 chains, each with 16667 iterations (first 2000 discarded), n.thin = 10 n.sims = 4401 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff delta -27.591 0.459 -28.494 -27.897 -27.593 -27.284 -26.682 1.001 4400 mu1 105.309 0.335 104.661 105.086 105.310 105.538 105.964 1.001 4400 mu2 77.718 0.314 77.098 77.506 77.713 77.925 78.329 1.001 4400 sigma1 2.598 0.248 2.161 2.422 2.576 2.752 3.142 1.002 1200 sigma2 1.977 0.228 1.593 1.817 1.956 2.111 2.512 1.001 3400 deviance 451.941 2.877 448.342 449.802 451.314 453.390 459.112 1.001 4400 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 4.1 and DIC = 456.1 DIC is an estimate of expected predictive error (lower deviance is better).
