Tutorial 15.1 - Multidimensional Scaling (MDS)
12 Mar 2015
> library(vegan) > library(ggplot2) > library(grid) > #define my common ggplot options > murray_opts <- opts(panel.grid.major=theme_blank(), + panel.grid.minor=theme_blank(), + panel.border = theme_blank(), + panel.background = theme_blank(), + axis.title.y=theme_text(size=15, vjust=0,angle=90), + axis.text.y=theme_text(size=12), + axis.title.x=theme_text(size=15, vjust=-1), + axis.text.x=theme_text(size=12), + axis.line = theme_segment(), + plot.margin=unit(c(0.5,0.5,1,2),"lines") + )
Error: Use 'theme' instead. (Defunct; last used in version 0.9.1)
> coenocline <- function(x,A0,m,r,a,g, int=T, noise=T) { + #x is the environmental range + #A0 is the maximum abundance of the species at the optimum environmental conditions + #m is the value of the environmental gradient that represents the optimum conditions for the species + #r the species range over the environmental gradient (niche width) + #a and g are shape parameters representing the skewness and kurtosis + # when a=g, the distribution is symmetrical + # when a>g - negative skew (large left tail) + # when a<g - positive skew (large right tail) + #int - indicates whether the responses should be rounded to integers (=T) + #noise - indicates whether or not random noise should be added (reflecting random sampling) + #NOTE. negative numbers converted to 0 + b <- a/(a+g) + d <- (b^a)*(1-b)^g + cc <- (A0/d)*((((x-m)/r)+b)^a)*((1-(((x-m)/r)+b))^g) + if (noise) {n <- A0/10; n[n<0]<-0; cc<-cc+rnorm(length(cc),0,n)} + cc[cc<0] <- 0 + cc[is.na(cc)]<-0 + if (int) cc<-round(cc,0) + cc + } > #plot(coenocline(0:100,40,40,20,1,1, int=T, noise=T), ylim=c(0,100))
To assist with demonstrating Multidimensional Scaling (MDS), we will return to the fabricated species abundance data introduced in Tutorial 13.2. This data set comprises the abundances of 10 species within 10 sites located along a transect that extends in a northerly direction over a mountain range.
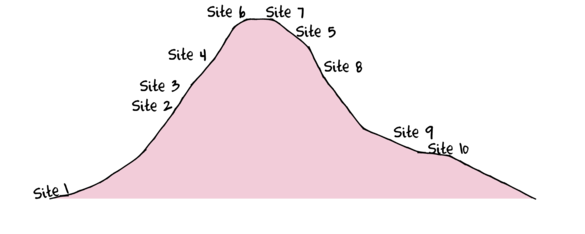
> set.seed(1) > x <- seq(0,50,l=10) > n <- 10 > sp1<-coenocline(x=x,A0=5,m=0,r=2,a=1,g=1,int=T, noise=T) > sp2<-coenocline(x=x,A0=70,m=7,r=30,a=1,g=1,int=T, noise=T) > sp3<-coenocline(x=x,A0=50,m=15,r=30,a=1,g=1,int=T, noise=T) > sp4<-coenocline(x=x,A0=7,m=25,r=20,a=0.4,g=0.1,int=T, noise=T) > sp5<-coenocline(x=x,A0=40,m=30,r=30,a=0.6,g=0.5,int=T, noise=T) > sp6<-coenocline(x=x,A0=15,m=35,r=15,a=0.2,g=0.3,int=T, noise=T) > sp7<-coenocline(x=x,A0=20,m=45,r=25,a=0.5,g=0.9,int=T, noise=T) > sp8<-coenocline(x=x,A0=5,m=45,r=5,a=1,g=1,int=T, noise=T) > sp9<-coenocline(x=x,A0=20,m=45,r=15,a=1,g=1,int=T, noise=T) > sp10<-coenocline(x=x,A0=30,m=50,r=5,a=1,g=1,int=T, noise=T) > X <- cbind(sp1, sp10,sp9,sp2,sp3,sp8,sp4,sp5,sp7,sp6) > #X<-X[c(1,10,9,2,3,8,4,5,7,6),] > colnames(X) <- paste("Sp",1:10,sep="") > rownames(X) <- paste("Site", c(1,10,9,2,3,8,4,5,7,6), sep="") > X <- X[c(1,4,5,7,8,10,9,6,3,2),] > data <- data.frame(Sites=factor(rownames(X),levels=rownames(X)), X)
| Sites | Sp1 | Sp2 | Sp3 | Sp4 | Sp5 | Sp6 | Sp7 | Sp8 | Sp9 | Sp10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Site1 | 5 | 0 | 0 | 65 | 5 | 0 | 0 | 0 | 0 | 0 |
| Site2 | 0 | 0 | 0 | 25 | 39 | 0 | 6 | 23 | 0 | 0 |
| Site3 | 0 | 0 | 0 | 6 | 42 | 0 | 6 | 31 | 0 | 0 |
| Site4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 40 | 0 | 14 |
| Site5 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 34 | 18 | 12 |
| Site6 | 0 | 29 | 12 | 0 | 0 | 0 | 0 | 0 | 22 | 0 |
| Site7 | 0 | 0 | 21 | 0 | 0 | 5 | 0 | 0 | 20 | 0 |
| Site8 | 0 | 0 | 0 | 0 | 13 | 0 | 6 | 37 | 0 | 0 |
| Site9 | 0 | 0 | 0 | 60 | 47 | 0 | 4 | 0 | 0 | 0 |
| Site10 | 0 | 0 | 0 | 72 | 34 | 0 | 0 | 0 | 0 | 0 |
Q-mode analyses
In contrast to R-mode analyses (such as principle components analysis), Q-mode analyses explore the patterns between objects (sites) on the basis of pairwise object likenesses (c.f. associations between variables). Whereas the starting point of R-mode analyses is typically either a covariance or correlation matrix and matrix algebra is used to arrange objects (sites) so as to preserve either their euclidean distances (PCA/RDA) or $\chi^2$ distances (CA/CCA), Q-mode analyses (such as MDS) provide euclidean or arbitrary (where only the order is preserved) ordination distances from any distance or similarity matrix.
This unrestricted choice of starting distance matrix provides enormous flexibility to be able to handle a wide range of data and issues. The distance matrix is selected to best reflect the nature of the data and thus acts as a sort of filter or conduit through which data are converted into a form suitable for analysis.
A consequence of basing analyses on distance matrices is that the resulting axes scores (coordinates) or patterns are not independent of one another. That is, the new values along a particular axes are all non-independent. Consequently, the new variables (dimensions) cannot be used in traditional linear modelling (c.f. PCA).
Multidimensional Scaling
The main function of multidimensional Scaling (MDS) is to re-project the objects (sites) in reduced dimension ordination space. As stated above, the axes scores cannot be used in subsequent analyses due to a lack of independence.
The real advantage of MDS over other ordination techniques is that it will generally represent the ordering relationships amongst objects within a given set of dimensions better than eigen-based ordinations. Whilst the eigen-based analyses rotates all of the original axes such that each axis maximizes the variance preserved by that axes, MDS constructs a configuration of objects specifically for only a nominated number of new axes. Therefore, when a 2 dimensional ordination is desired, the configuration from an MDS is optimized for just two dimensions whereas the eigen-based object configurations are independent of how many dimensions you intend to plot.
Principle components analysis (PCA) can be performed by either spectral (eigen) decomposition of an association matrix or single value decomposition of the original data matrix. Either way, it yields a rigid rotation of axes in that the positions of points relative to one another (euclidean distances) are maintained during rotation. The rotated axes are referred to as principle components.
Obviously, the higher the degree of associations between the variables (species), the more strongly focused and oriented the data cloud is and therefore the more 'successful' the PCA in terms of being able to represent many variables by only a few new variables.
The resulting principle components (axes) and scores (new variable values) are independent of one another and therefore the 'reduced data' can be used in subsequent statistical analyses. For example, principle component 1 (as one measure of the community) could be regressed against an environmental variable (such as altitude or rainfall) in a regular linear modelling procedure. Similarly, multiple correlated predictor variables (that otherwise violate multi-collinearity) could be reduced to one or two orthogonal predictors to use in linear modelling. Principle components analysis is therefore often an intermediate step to used as either responses or predictors in linear modeling.
MDS is mathematically and conceptually relatively simple, yet computationally very intense. Rather than have elegant mathematical solutions, MDS works on bruit grunt.
We will now attempt to conceptualize the process of multidimensional scaling
- Generate a distance (dissimiliarity) matrix from the multivariate data.
For the purpose of this demonstration (and since the data are species abundances), we will use a Bray-Curtis dissimilarity
matrix.
Note that this is a triangular matrix - the diagonals (which would all be 0) and the upper right cells (which are a mirror of the lower left cells) are omitted. For the remainder of this demonstration, we will represent this dissimilarity matrix as:
> data.dist <- vegdist(data[,-1], "bray") > data.dist
Site1 Site2 Site3 Site4 Site2 0.6429 Site3 0.8625 0.1685 Site4 1.0000 0.6871 0.5540 Site5 1.0000 0.7178 0.6000 0.2581 Site6 1.0000 1.0000 1.0000 1.0000 Site7 1.0000 1.0000 1.0000 1.0000 Site8 0.9237 0.4362 0.2908 0.3273 Site9 0.3011 0.3333 0.4694 1.0000 Site10 0.2265 0.4070 0.5812 1.0000 Site5 Site6 Site7 Site8 Site2 Site3 Site4 Site5 Site6 0.6391 Site7 0.5862 0.4128 Site8 0.4603 1.0000 1.0000 Site9 1.0000 1.0000 1.0000 0.7964 Site10 1.0000 1.0000 1.0000 0.8395 Site9 Site2 Site3 Site4 Site5 Site6 Site7 Site8 Site9 Site10 0.1336
- Decide on how many dimensions are required of the final ordination plot - how many dimensions do you want to visualize the objects (sites) in? Choice of the number of dimensions is typically a compromise between ease of visualization (fewer dimensions) and sufficiently capturing the patterns in the original data (more dimensions). Two or three dimensions is usually a decent compromise that is nonetheless based more on ease of visualization.
- Having nominated the number of ordination dimensions (in this case 2), we then randomly arrange our sites in two dimensions.
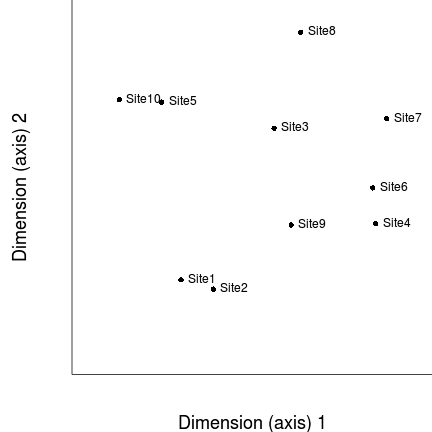
- We now measure how well this configuration (arrangement of sites on the ordination) match the original distance (dissimilarity) matrix.
That is, we measure how well the two new axes represent the patterns that existed across all the original variables (species).
We do this by creating a matrix of distances between all of the points on the ordination. These are called ordination distances and are
simply the euclidean (straight-line) distances between each point and each other point.
The degree of agreement between the data distance and ordination distance matrices is usually explored by fitting a regression model. The relationship can also be visualized in a Shepard diagram. The Shepard plots ordination distances (y-axis) against data distances (x-axis).
Obviously since the sites have just been placed in ordination space at random, there will initially be a very low degree of correspondence between the original data distance matrix and the new ordination distance matrix.
The goodness of fit of the regression between data and ordination distances is measured by a property called stress. There are basically two forms of stress:
- Kruskal's stress $$stress = \sqrt{\frac{\sum{(D_{ord.i}-D_{data.i})^2}}{\sum{D_{ord.i^2}}}}$$
- Modified Kruskal's stress $$stress = \sqrt{\frac{\sum{(D_{ord.i}-D_{data.i})^2}}{\sum{D_{ord.i^2}-\bar{D_{data.i}}}}}$$ The modified Kruskal's stress is equivalent to $\sqrt(1-R^2)$
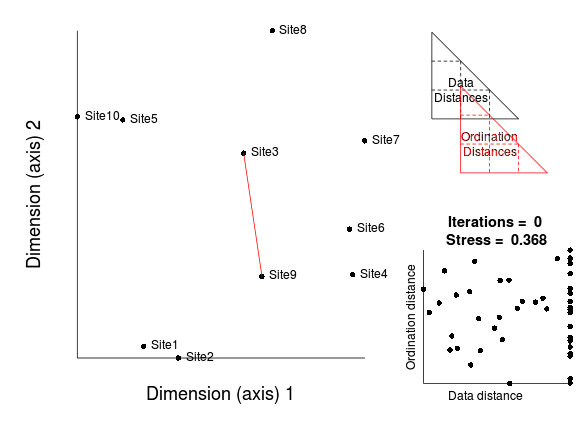
- The configuration of points on the ordination is then "improved" by shifting the position of the points
slightly. Points are shifted via a process called gradient descent which ensures that points are shifted in the
direction that will cause the stress value to be reduced by the greatest amount (the steepest descent).
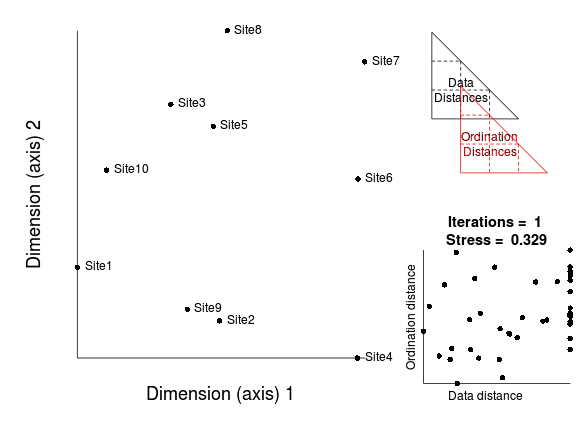
- The points are repeatedly (iteratively) adjusted. Each time the correlation between data and ordination distances will improve.
At first, successive iterations will yield marked improvements in the correlation between data and ordination distances (and thus large improvements in stress). However, eventually the improvements will be very minor.
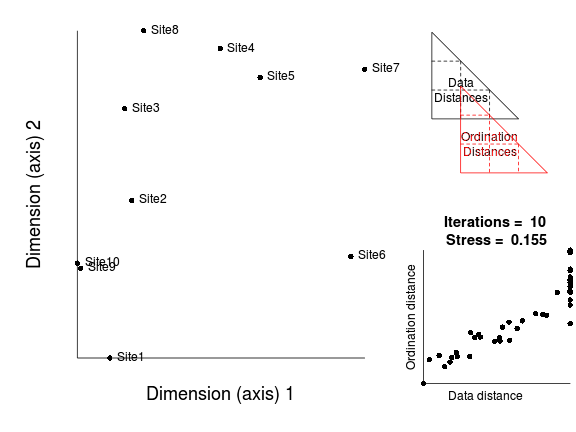 The iterative process could in theory go on indefinitely. However as the rate of improvement diminishes with each additional iteration, there comes a point were the improvements become imperceptible. There is also the possibility that there was no real patter in the original data and therefore no amount of exhaustive reconfiguration is going to yield patterns in reduced ordination space.
The iterative process could in theory go on indefinitely. However as the rate of improvement diminishes with each additional iteration, there comes a point were the improvements become imperceptible. There is also the possibility that there was no real patter in the original data and therefore no amount of exhaustive reconfiguration is going to yield patterns in reduced ordination space.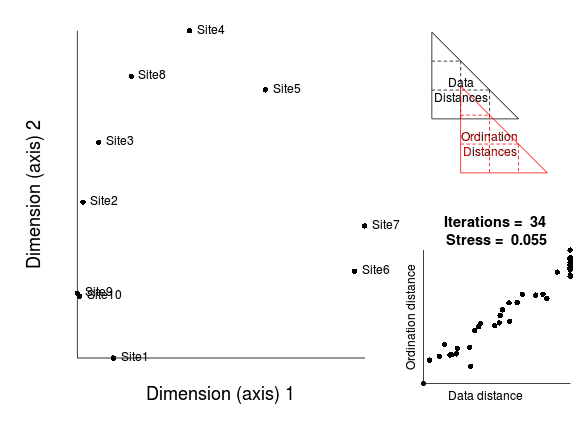
There are typically three stopping rules in place that indicate when the iterative reconfiguration process should cease.
- convergence tolerance - iterations stop when the stress value falls below a threshold (such as stress=0.001)
- the stress ratio (ratio of the stress values for the current and previous iterations) - iterations stop once the improvement in stress falls below a threshold (such as 1.1)
- maximum iterations - iterations stop once a set number of iterations have occurred (such as 200).
Ideally, we are seeking a stress value of less than 0.2. This roughly corresponds to the ordination distances (and thus the new axes from which they are derived) explaining 80% of the variability in the data dissimilarity matrix (which is based on all the original variables). Hence if we have a stress value of 0.2 or less, then we can be increasingly more confident that we have been able to encapsulate the major patterns.
The final configuration displays an arrangement of objects (sites) in two (or three) dimensions in which the rank proximity of points to one another is proportional to how alike the sites are in their community structure. Note however, the axes themselves have no real meaning. Unlike principle components analysis or correspondence analysis in which the axes reflect major underlying gradients, the orientation of points with respect to the MDS axes are somewhat arbitrary. Instead the axes operate together just to create the ordination space and the configuration can be oriented at any angle to the axes. That said, it is possible to lock the orientation of the configuration so as to give the individual axes more meaning.
Starting configuration
The starting configuration can (as demonstrated above) be any random arrangement of points in the nominated dimensions. Alternatively, the starting configuration can be approximated by first running an eigen-analysis based ordination (similar to a PCA, yet using a distance matrix rather than an association matrix). Not only does this usually reduce the risks of running into local minimum issues (see below), it can also drastically reduce the number of iteration required for the iterations to converge.
The final configuration can be dependent on the starting configuration (i.e. the initial arrangement of sites). The gradient descent algorithms can get stuck in what are called local minimums (or optimums). This occurs when the math limits the possible position adjustments that can be made and the small alterations possible within the scope of the algorithms are insufficient to bring about substantial improvements. In these cases, more drastic adjustments would be required in order to free up the algorithm to find true minimums (better configurations).
Therefore, it is usually good practice to repeat the entire process multiple times, each time with a different random starting configuration.
Procrustes rotation
The final stress value of a final configuration is a good indicator of how good the configuration is. We could therefore, run the analysis multiple times (each time with a different starting configuration) and ultimately select the final configuration that has the lowest stress value. However, this might lead to many extraneous runs. It may be that the data and patterns are sufficiently strong and stable that any two runs would result in essentially the same configuration (irrespective of initial configuration). What is required is a means to evaluate when the final configurations of two runs are essentially the same (when they have converged on the same solution).
Unlike eigen-based analysis (such as PCA) in which the first axis represents the orientation of data in the plane of greatest variance and each subsequent axis represents the greatest direction of remaining variance, in MDS, the orientation and scaling of points with respect to the axes are of no meaning. The two (or three) axes simply define the ordination space in which the point can be plotted. Consequently it can often be difficult to judge how similar two configurations are - they might be identical, yet one is upside down and twice as wide (for example).
Consider the following two final configurations resulting from the same dissimilarity matrix and with the same number of iterations and differing only in their starting configurations.
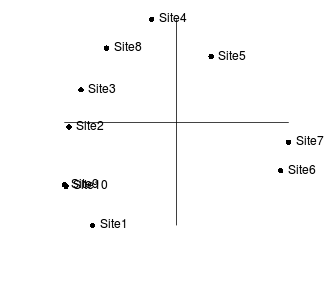 |
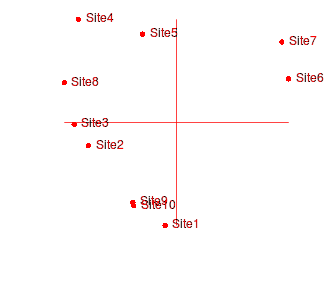 |
If we rotated the configuration on the right clockwise approximately 45 degrees, these points would superimpose almost exactly onto the configuration on the left. This can be seen in the figure below (left). The previous best configuration is depicted in solid black and the latest configuration is depicted in light red and solid red when scaled and rotated and superimposed. Blue arrows indicated the amount that each point is transformed. The figure on the right indicates the differences (residuals) between the two configurations (once projection on the same scale and rotation) as solid red lines.
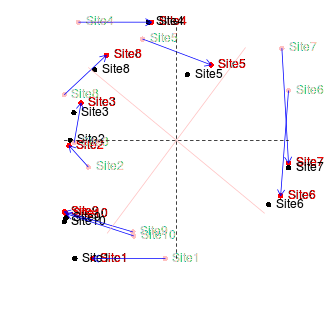 |
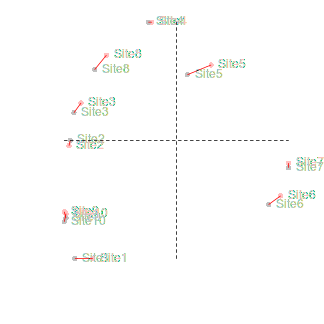 |
This process of comparing the two final configurations by scaling and rotating one configuration (the latest configuration) and superimposing it upon another (the previous best configuration - the standard) is called Procrustes rotation. The amount of scaling and rotation is selected so as to minimize the sum of the squared distances (residuals - the red lines in the figure above) between the two sets of points (analogous to the way a regression line is fitted such that the squared deviations from the line are minimized). This minimum sum of squares is called the root mean square error (rmse). The smaller the rmse, the more similar the two configurations are.
Two final configurations are considered to have converged (arrived at essentially the same solution) when the rmse is less than 0.01, and no single residual value exceeds 0.005. Procrustes analysis thereby provides a mechanism for determining when to stop repeatedly re-running the analysis - stop when there is convergence as measured by procrustes rmse.
MDS options
There is not just one single multidimensional scaling process. Rather, there are many different process and options, each of which alters some aspect of the behavior of the MDS. Here are some of the more major choices:
- metric versus non-metric. This is a fundamental distinction between those MDS's that base stress (and therefore the stopping criterion) on
metric regression (essentially linear regression) and those that base stress on monotonic (essentially non-linear) regression. For most
ecological data, the relationship between data dissimilarity and ordination distance will be non-linear. Consequently, non-metric multidimensional scaling (NMDS)
is preferred by ecologists.
Within NMDS, there is a further distinction between whether the regression should be:
- monotonic regression
- or an equivalent of a locally weighted regression separate for each point
- how to generate the initial or starting configuration. The MDS can either start with a completely random configuration or the configuration can be leveraged from other means.
- whether or not to automatically employ multiple random starts.
- whether an how to scale the axes scores. This can make the ordinations easier to interpret.
In R, MDS is implemented several functions (although I will only mention two per method).
- metric MDS is performed by either cmdscale() or metaMDS() (vegan)
- non-metric MDS is performed by either isoMDS() or metaMDS() (vegan).
metaMDS()
The metaMDS() function does not itself perform multidimensional scaling. Rather it is a short-cut wrapper that brings together each of the important steps in performing multidimensional scaling.
- If the metaMDS() function is provided raw community data, it will:
- transform and/or standardize the data. Depending on the magnitude of the abundances, data will be square-root transformed before being subject to a Wisconsin double standardization (column maximum followed by row total). This behavior can be suppressed via the autotransform=FALSE argument to allow full control over transformations and standardizations.
- generate a dissimilarity matrix. The vegdist() function is called to generate a dissimilarity matrix. By default this is a Bray-Curtis dissimilarity, however, any of the distances supported by vegdist() can be nominated.
- The above steps are skipped if the metaMDS() function is provided with a dissimilarity matrix (matrix of class dist) instead of a raw data matrix.
- The MDS engine is called. By default this is the monoMDS() function also in the vegan package, although it is also possible to
specify isoMDS() as an alternative.
-
The initial starting configuration (if not provided) is generated via metric scaling (principle coordinates analysis - PCoA).
PCoA is essentially a PCA performed on a distance matrix.
Thereafter, the monoMDS() engine function is called repeatedly (up to trymax=20 times), each time with a new completely random starting configuration. Each time monoMDS is called, the engine iteratively adjusts the positions of points on the ordination space and evaluates the stress with respect to the stopping criterion.
- The final stress value from each new run of the monoMDS() function is compared to the final stress value from the previous 'best' run and if the new stress value is better (lower), then the final configurations of the previous best and the latest runs are compared via Procrustes analysis to establish whether they might have converged (essentially arrived at the same configuration). If they have converged, the configuration with the lower stress value is considered the 'best' configuration.
-
The initial starting configuration (if not provided) is generated via metric scaling (principle coordinates analysis - PCoA).
PCoA is essentially a PCA performed on a distance matrix.
- Following the arrival of the final configuration, the results are scaled to allow:
- center the origin (0,0) to the average of the axes scores
- either principle components-like rotation of the configuration so as to mimic the way the first axis of a PCA represents the orientation of the greatest variance or rotating the configuration such that the first axis (dimension) is parallel to an environmental variable.
- Finally, variable (species) axes scores are generated as the weighted averages.
Our example data set is species abundance data, and therefore the defaults (Wisconsin double standardization, Bray-Curtis dissimilarity, non-metric multidimensional scaling with multiple random starts followed by centering and principle components-like rotation of the configuration, would seem appropriate.
> library(vegan) > data.nmds <- metaMDS(data[,-1])
Square root transformation Wisconsin double standardization Run 0 stress 0.03499 Run 1 stress 9.419e-05 ... New best solution ... procrustes: rmse 0.115 max resid 0.2088 Run 2 stress 0.0005712 ... procrustes: rmse 0.1885 max resid 0.2961 Run 3 stress 0.0004598 ... procrustes: rmse 0.1281 max resid 0.2567 Run 4 stress 0.1072 Run 5 stress 0.08642 Run 6 stress 0.000249 ... procrustes: rmse 0.1767 max resid 0.2933 Run 7 stress 9.78e-05 ... procrustes: rmse 0.1051 max resid 0.2432 Run 8 stress 9.692e-05 ... procrustes: rmse 0.1516 max resid 0.3103 Run 9 stress 9.796e-05 ... procrustes: rmse 0.1283 max resid 0.2929 Run 10 stress 9.757e-05 ... procrustes: rmse 0.06026 max resid 0.0896 Run 11 stress 9.905e-05 ... procrustes: rmse 0.1624 max resid 0.3806 Run 12 stress 0.07878 Run 13 stress 9.678e-05 ... procrustes: rmse 0.008004 max resid 0.01477 Run 14 stress 0.001213 Run 15 stress 0.002591 Run 16 stress 9.385e-05 ... New best solution ... procrustes: rmse 0.1073 max resid 0.2635 Run 17 stress 9.784e-05 ... procrustes: rmse 0.1069 max resid 0.2275 Run 18 stress 0.00139 Run 19 stress 9.781e-05 ... procrustes: rmse 0.02273 max resid 0.04333 Run 20 stress 9.553e-05 ... procrustes: rmse 0.1389 max resid 0.2901
- the original data was subject to square-root transformation followed by a Wisconsin double standardization.
- a total of 20 random starts (default) were attempted. For each run, the final stress is given and when a stress value similar to the lowest is achieved, a procrustes analysis determines the rmse and maximum residual value (to determine whether convergence has occurred).
- The rmse value did not fall bellow 0.01 and therefore convergence was not considered to have occurred prior to the completion of the set number of random starts.
Additional information is gained by printing the object.
> data.nmds
Call: metaMDS(comm = data[, -1]) global Multidimensional Scaling using monoMDS Data: wisconsin(sqrt(data[, -1])) Distance: bray Dimensions: 2 Stress: 9.385e-05 Stress type 1, weak ties No convergent solutions - best solution after 20 tries Scaling: centring, PC rotation, halfchange scaling Species: expanded scores based on 'wisconsin(sqrt(data[, -1]))'
- we are reminded that the configuration is for two dimensions
- the final stress of the 'best' configuration is indicated
- we are reminded of any scaling and rotations that might have been applied to the axes scores. In this case, the scores were centered and rotated to emulate the way PCA orients axes according to planes of greatest variance.
- we are informed that variable (species) scores have also been calculated as the means of each of the site scores weighted by the square-root species abundances.
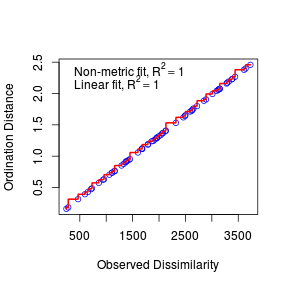
Lets examine the Shepard (stress) plot to get a feel for how well the configuration in two dimensions matches the original data patterns.
> stressplot(data.nmds)
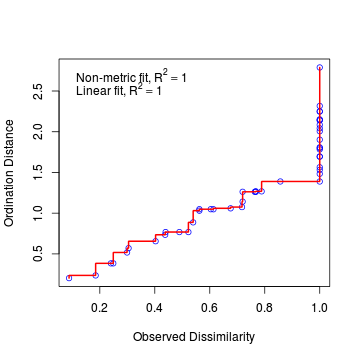
- The fit between the community distances (data dissimilarity) and ordination distances is indicated by the monotonic step line (red).
- Two stress values are provided:
- Non-metric fit - this is the modified Kruskal's stress value and is $\sqrt{1-R^2}$
- Linear fit - this is the $R^2$ value of the correlation between the ordination values and the ordination values predicted from the (monotonic) regression line.
> ordiplot(data.nmds, type="text")
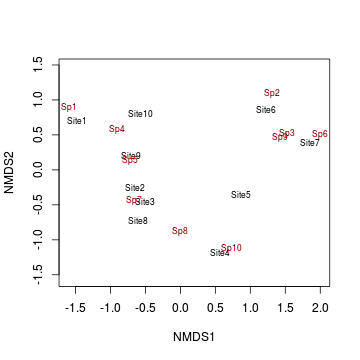
Interpreting axes in terms of environmental variables
Establishing relationships between MDS axes and environmental variables is not as straight forward as it is in PCA. Firstly, the orientation of sites in MDS reduced ordination space is largely arbitrary. Thus, in order to relate them to changes in environmental factors, the only sensible way to orient the axes is in accordance to planes of greatest variability (similar to PCA). So the axes scores must first be rotated accordingly.
Secondly, as mentioned previously, as the axes scores are based on distance measures (which are themselves derived from the patterns between objects (sites), they scores are not independent of one another. Consequently, they violate one of the main assumptions of inferential statistics.
Nevertheless, environmental variables can be overlayed onto the ordination plot in a similar way as the species scores are overlayed. Lets first retrieve the environmental variables collected from the 10 sites.
> set.seed(1) > Site <- gl(10,1,10,lab=paste('Site',1:10, sep="")) > Y <- matrix(c( + 6.1,4.2,101325,2, + 6.7,9.2,101352,510, + 6.8,8.6,101356,546, + 7.0,7.4,101372,758, + 7.2,5.8,101384,813, + 7.5,8.4,101395,856, + 7.5,0.5,101396,854, + 7.0,11.8,101370,734, + 8.4,8.2,101347,360, + 6.2,1.5,101345,356 + ),10,4, byrow=TRUE) > colnames(Y) <- c('pH','Slope', 'Pressure', 'Altitude') > Substrate <- factor(c('Quartz','Shale','Shale','Shale','Shale','Quartz','Quartz','Shale','Quartz','Quartz')) > enviro <- data.frame(Site,Y,Substrate)
> ordiplot(data.nmds, type="text") > data.envfit <- envfit(data.nmds,enviro[,-1]) > plot(data.envfit)
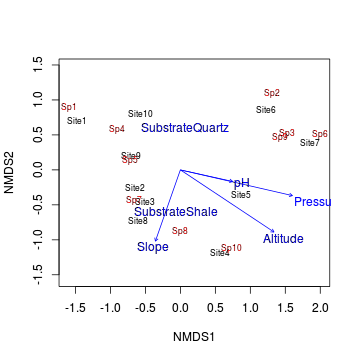
- Altitude, pressure and pH all seem to align predominantly with axis 1 (particularly pressure)
- Temperature and slope align almost entirely with axis 2.
- Arrow length is proportional to the degree of correlation between the environmental variable and the ordination
The envfit() function performs one such permutation test and is based on comparing the observed $R^2$ to a large number of $R^2$ values from repeatedly shuffling the rows of the environmental data.
> data.envfit
***VECTORS
NMDS1 NMDS2 r2 Pr(>r)
pH 0.976 -0.217 0.20 0.471
Slope -0.331 -0.944 0.39 0.176
Pressure 0.975 -0.224 0.90 0.001 ***
Altitude 0.833 -0.553 0.86 0.001 ***
---
Signif. codes:
0 '***' 0.001 '**' 0.01 '*' 0.05
'.' 0.1 ' ' 1
P values based on 999 permutations.
***FACTORS:
Centroids:
NMDS1 NMDS2
SubstrateQuartz 0.06 0.59
SubstrateShale -0.06 -0.59
Goodness of fit:
r2 Pr(>r)
Substrate 0.25 0.11
P values based on 999 permutations.
- For each of the environmental variables the table displays:
- the cosine of the angle to each axis
- the $R^2$ of the relationship between the environmental variable and the ordination
- permutation-based p-value
- The chances of obtaining the relationships between ordination and Temperature, Pressure and Altitude by chance are all very low ($<0.05$), hence we conclude that Temperature, Pressure and Altitude are all correlated to community structure.
There are numerous alternative ways of performing permutation test (based on other test statistics and randomization schedules), some of which will be explored in Tutorial 15.2.
Worked Examples
Basic statistics references
- Legendre and Legendre
- Quinn & Keough (2002) - Chpt 17
Non-metric Multidimensional Scaling (NMDS)
The following example is designed to help you appreciate the link between distance measures and ordination space (MDS). The data set consists of distances (km) between major Australia cities (as the crow flies), and is in the form of a triangular matrix.
Download austcities data set| Format of austcites.csv data file | |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|||||||||||||||||||||||||||||||||||
 |
> austcities <- read.csv('../downloads/data/austcities.csv') > austcities
Canberra Sydney Melbourne
Canberra 0 NA NA
Sydney 246 0 NA
Melbourne 467 713 0
Adelaide 958 1160 653
Perth 3090 3290 2720
Broome 3280 3380 3120
Alice 1950 2030 1890
Darwin 3130 3150 3150
Cairns 2060 1960 2320
Townsville 1790 1680 2070
Brisbane 941 731 1370
Hobart 860 1060 597
Adelaide Perth Broome Alice
Canberra NA NA NA NA
Sydney NA NA NA NA
Melbourne NA NA NA NA
Adelaide 0 NA NA NA
Perth 2130 0 NA NA
Broome 2490 1680 0 NA
Alice 1330 1990 1370 0
Darwin 2620 2650 1110 1290
Cairns 2120 3440 2500 1450
Townsville 1920 3390 2590 1420
Brisbane 1600 3610 3320 1960
Hobart 1160 3010 3640 2460
Darwin Cairns Townsville
Canberra NA NA NA
Sydney NA NA NA
Melbourne NA NA NA
Adelaide NA NA NA
Perth NA NA NA
Broome NA NA NA
Alice NA NA NA
Darwin 0 NA NA
Cairns 1680 0 NA
Townsville 1860 281 0
Brisbane 2850 1390 1110
Hobart 3730 2890 2630
Brisbane Hobart
Canberra NA NA
Sydney NA NA
Melbourne NA NA
Adelaide NA NA
Perth NA NA
Broome NA NA
Alice NA NA
Darwin NA NA
Cairns NA NA
Townsville NA NA
Brisbane 0 NA
Hobart 1790 0
Note the format of the file, it is a triangular distance matrix.
- While the file is a distance matrix, at this stage R is unaware of it, we must manually make it aware (a round about way of saying that we must type a command to force R to treat the data set as a distance matrix.
Convert the data frame into a distance matrix (HINT).
Show code
> austcities <- as.dist(austcities) > austcities
Canberra Sydney Melbourne Sydney 246 Melbourne 467 713 Adelaide 958 1160 653 Perth 3090 3290 2720 Broome 3280 3380 3120 Alice 1950 2030 1890 Darwin 3130 3150 3150 Cairns 2060 1960 2320 Townsville 1790 1680 2070 Brisbane 941 731 1370 Hobart 860 1060 597 Adelaide Perth Broome Alice Sydney Melbourne Adelaide Perth 2130 Broome 2490 1680 Alice 1330 1990 1370 Darwin 2620 2650 1110 1290 Cairns 2120 3440 2500 1450 Townsville 1920 3390 2590 1420 Brisbane 1600 3610 3320 1960 Hobart 1160 3010 3640 2460 Darwin Cairns Townsville Sydney Melbourne Adelaide Perth Broome Alice Darwin Cairns 1680 Townsville 1860 281 Brisbane 2850 1390 1110 Hobart 3730 2890 2630 Brisbane Sydney Melbourne Adelaide Perth Broome Alice Darwin Cairns Townsville Brisbane Hobart 1790 -
Perform an MDS with 2 dimensions on the city distances matrix (HINT).
Show code
> library(vegan) > austcities.mds <- metaMDS(austcities, k=2)
Run 0 stress 0 Run 1 stress 0 ... procrustes: rmse 0.004977 max resid 0.01025 Run 2 stress 2.278e-05 ... procrustes: rmse 0.004939 max resid 0.00979 *** Solution reached
- What was the final stress value (as a percentage)?
Show code
> austcities.mds$stress
[1] 0
- What does this stress value suggest about the success of the MDS?
- Generate a Shepard diagram.
The Shepard diagram (plot) represents the relationship between the original distances (y-axis) and the new MDS ordination distances (x-axis).
Does this and the stress value indicate that the patterns present in the original distance matrix (crow flies distances between cities)
are adequately reproduced from the 2 new dimensions?
Show code
> stressplot(austcities.mds)
- What was the final stress value (as a percentage)?
-
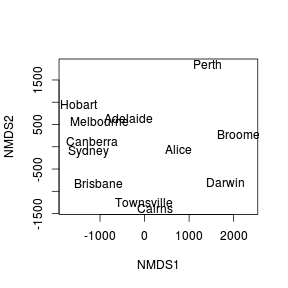
Generate an ordination plot.
The final ordination plot summarizes the relationship between the cities.
Does this ordination plot approximate the true geographical arrangement of the cities?
Show codeRecall that the orientation of points in a MDS ordination plot is arbitrary. Unlike PCA and CA, the points are not orientated towards axes explaining most variance. Instead the two new dimensions simply provide the space or scope in which the points are arranged. What if we rotated the points 140 degrees. Does this make the patterns easier to relate back to reality?
> ordiplot(austcities.mds, display="sites", type="n") > text(austcities.mds,lab=rownames(austcities.mds$points))
 Show code
Show code> library(shape) > austcities.rot <-rotatexy(austcities.mds$points, angle=140) > plot(austcities.rot, type="n") > text(austcities.rot,lab=rownames(austcities.mds$points))
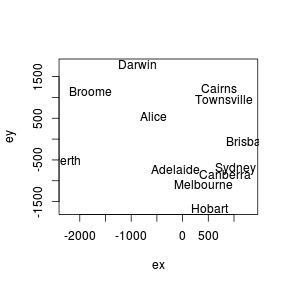
- In this case, what might the two new MDS dimensions (variables) represent?
(hint think of the ordination plot as a map)
We are now ready to perform the MDS for the purpose of examining the ordination plot.
Non-metric Multidimensional Scaling (NMDS)
Mac Nally (1989) studied geographic variation in forest bird communities. His data set consists of the maximum abundance for 102 bird species from 37 sites that where further classified into five different forest types (Gippsland manna gum, montane forest, woodland, box-ironbark and river redgum and mixed forest). He was primarily interested in determining whether the bird assemblages differed between forest types.
Download the macnally data set| Format of macnally_full.csv data file | |||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
 |
> macnally <- read.csv('../downloads/data/macnally_full.csv') > macnally
HABITAT GST
Reedy Lake Mixed 3.4
Pearcedale Gipps.Manna 3.4
Warneet Gipps.Manna 8.4
Cranbourne Gipps.Manna 3.0
Lysterfield Mixed 5.6
Red Hill Mixed 8.1
Devilbend Mixed 8.3
Olinda Mixed 4.6
Fern Tree Gum Montane Forest 3.2
Sherwin Foothills Woodland 4.6
Heathcote Ju Montane Forest 3.7
Warburton Montane Forest 3.8
Millgrove Mixed 5.4
Ben Cairn Mixed 3.1
Panton Gap Montane Forest 3.8
OShannassy Mixed 9.6
Ghin Ghin Mixed 3.4
Minto Mixed 5.6
Hawke Mixed 1.7
St Andrews Foothills Woodland 4.7
Nepean Foothills Woodland 14.0
Cape Schanck Mixed 6.0
Balnarring Mixed 4.1
Bittern Gipps.Manna 6.5
Bailieston Box-Ironbark 6.5
Donna Buang Mixed 1.5
Upper Yarra Mixed 4.7
Gembrook Mixed 7.5
Arcadia River Red Gum 3.1
Undera River Red Gum 2.7
Coomboona River Red Gum 4.4
Toolamba River Red Gum 3.0
Rushworth Box-Ironbark 2.1
Sayers Box-Ironbark 2.6
Waranga Mixed 3.0
Costerfield Box-Ironbark 7.1
Tallarook Foothills Woodland 4.3
EYR GF BTH GWH WTTR
Reedy Lake 0.0 0.0 0.0 0.0 0.0
Pearcedale 9.2 0.0 0.0 0.0 0.0
Warneet 3.8 0.7 2.8 0.0 0.0
Cranbourne 5.0 0.0 5.0 2.0 0.0
Lysterfield 5.6 12.9 12.2 9.5 2.1
Red Hill 4.1 10.9 24.5 5.6 6.7
Devilbend 7.1 6.9 29.1 4.2 2.0
Olinda 5.3 11.1 28.2 3.9 6.5
Fern Tree Gum 5.2 8.3 18.2 3.8 4.2
Sherwin 1.2 4.6 6.5 2.3 5.2
Heathcote Ju 2.5 6.3 24.9 2.8 7.4
Warburton 6.5 11.1 36.1 6.2 8.5
Millgrove 6.5 11.9 19.6 3.3 8.6
Ben Cairn 9.3 11.1 25.9 9.3 8.3
Panton Gap 3.8 10.3 34.6 7.9 4.8
OShannassy 4.0 5.4 34.9 7.0 5.1
Ghin Ghin 2.7 9.1 16.1 1.3 3.2
Minto 3.3 13.3 28.0 7.0 8.3
Hawke 2.6 5.5 16.0 4.3 6.7
St Andrews 3.6 6.0 25.2 3.7 7.5
Nepean 5.6 5.5 20.0 3.0 6.6
Cape Schanck 4.9 4.9 16.2 3.4 2.6
Balnarring 4.9 10.7 21.2 3.9 0.0
Bittern 9.7 7.8 14.4 5.2 0.0
Bailieston 2.5 5.1 5.6 4.3 5.7
Donna Buang 0.0 2.2 9.6 6.7 3.0
Upper Yarra 3.1 7.0 17.1 8.3 12.8
Gembrook 7.5 12.7 16.4 4.7 6.4
Arcadia 0.0 1.2 0.0 1.2 0.0
Undera 0.0 2.2 0.0 1.3 6.5
Coomboona 0.0 2.1 0.0 0.0 3.3
Toolamba 0.0 0.5 0.0 0.8 0.0
Rushworth 1.1 3.2 1.8 0.5 4.8
Sayers 0.0 1.1 7.5 1.6 5.2
Waranga 1.6 1.5 3.0 0.0 3.0
Costerfield 2.2 4.5 9.0 2.7 6.0
Tallarook 2.9 8.7 14.4 2.9 5.8
WEHE WNHE SFW WBSW CR
Reedy Lake 0.0 11.9 0.4 0.0 1.1
Pearcedale 0.0 11.5 8.3 12.6 0.0
Warneet 10.7 12.3 4.9 10.7 0.0
Cranbourne 3.0 10.0 6.9 12.0 0.0
Lysterfield 7.9 28.6 9.2 5.0 19.1
Red Hill 9.4 6.7 0.0 8.9 12.1
Devilbend 7.1 27.4 13.1 2.8 0.0
Olinda 2.6 10.9 3.1 8.6 9.3
Fern Tree Gum 2.8 9.0 3.8 5.6 14.1
Sherwin 0.6 3.6 3.8 3.0 7.5
Heathcote Ju 1.3 4.7 5.5 9.5 5.7
Warburton 2.3 25.4 8.2 5.9 10.5
Millgrove 2.5 11.9 4.3 5.4 10.8
Ben Cairn 2.8 2.8 2.8 8.3 18.5
Panton Gap 2.9 3.7 4.8 7.2 5.9
OShannassy 2.6 6.4 3.9 11.3 11.6
Ghin Ghin 4.7 0.0 22.0 5.8 7.4
Minto 7.0 38.9 10.5 7.0 14.0
Hawke 3.5 5.9 6.7 10.0 3.7
St Andrews 4.7 10.0 0.0 0.0 4.0
Nepean 7.0 3.3 7.0 10.0 4.7
Cape Schanck 2.8 9.4 6.6 7.8 5.1
Balnarring 5.1 2.9 12.1 6.1 0.0
Bittern 11.5 12.5 20.7 4.9 0.0
Bailieston 6.2 6.2 1.2 0.0 0.0
Donna Buang 8.1 0.0 0.0 7.3 8.1
Upper Yarra 1.3 6.4 2.3 5.4 5.4
Gembrook 1.6 8.9 9.3 6.4 4.8
Arcadia 0.0 1.8 0.7 0.0 0.0
Undera 0.0 0.0 6.5 0.0 0.0
Coomboona 0.0 0.0 0.8 0.0 0.0
Toolamba 0.0 0.0 1.6 0.0 0.0
Rushworth 0.9 5.3 4.8 0.0 1.1
Sayers 3.6 6.9 6.7 0.0 2.7
Waranga 0.0 14.5 6.7 0.0 0.7
Costerfield 2.5 7.7 9.5 0.0 7.7
Tallarook 2.8 11.1 2.9 0.0 3.8
LK RWB AUR STTH LR
Reedy Lake 3.8 9.7 0.0 0.0 4.8
Pearcedale 0.5 11.6 0.0 0.0 3.7
Warneet 1.9 16.6 2.3 2.8 5.5
Cranbourne 2.0 11.0 1.5 0.0 11.0
Lysterfield 3.6 5.7 8.8 7.0 1.6
Red Hill 6.7 2.7 0.0 16.8 3.4
Devilbend 2.8 2.4 2.8 13.9 0.0
Olinda 3.8 0.6 1.3 10.2 0.0
Fern Tree Gum 3.2 0.0 0.0 12.2 0.6
Sherwin 2.4 0.6 0.0 11.3 5.8
Heathcote Ju 2.9 0.0 1.8 12.0 0.0
Warburton 3.1 9.8 1.6 7.6 15.0
Millgrove 6.5 2.7 2.0 8.6 0.0
Ben Cairn 3.1 0.0 3.1 12.0 3.3
Panton Gap 3.1 0.6 3.8 17.3 2.4
OShannassy 2.3 0.0 2.3 7.8 0.0
Ghin Ghin 4.5 0.0 0.0 8.1 2.7
Minto 5.2 1.7 5.2 25.2 0.0
Hawke 2.1 0.5 1.5 9.0 4.8
St Andrews 5.1 2.8 3.7 15.8 3.4
Nepean 3.3 2.1 3.7 12.0 2.2
Cape Schanck 5.2 21.3 0.0 0.0 4.3
Balnarring 2.7 0.0 0.0 4.9 16.5
Bittern 0.0 16.1 5.2 0.0 0.0
Bailieston 1.6 5.0 4.1 9.8 0.0
Donna Buang 1.5 2.2 0.7 5.2 0.0
Upper Yarra 2.4 0.6 2.3 6.4 0.0
Gembrook 3.6 14.5 4.7 24.3 2.4
Arcadia 1.8 0.0 2.5 0.0 2.7
Undera 0.0 0.0 2.2 7.5 3.1
Coomboona 2.8 0.0 2.2 3.1 1.7
Toolamba 2.0 0.0 2.5 0.0 2.5
Rushworth 1.1 26.3 1.6 3.2 0.0
Sayers 1.6 8.0 1.6 7.5 2.7
Waranga 4.0 23.0 1.6 0.0 8.9
Costerfield 2.2 8.9 1.9 9.3 1.1
Tallarook 2.9 2.9 1.9 4.6 10.3
WPHE YTH ER PCU ESP
Reedy Lake 27.3 0.0 5.1 0.0 0.0
Pearcedale 27.6 0.0 2.7 0.0 3.7
Warneet 27.5 0.0 5.3 0.0 0.0
Cranbourne 20.0 0.0 2.1 0.0 2.0
Lysterfield 0.0 0.0 1.4 0.0 3.5
Red Hill 0.0 0.0 2.2 0.0 3.4
Devilbend 16.7 0.0 0.0 0.0 5.5
Olinda 0.0 0.0 1.2 0.0 5.1
Fern Tree Gum 0.0 0.0 1.3 2.8 7.1
Sherwin 0.0 9.6 2.3 2.9 0.6
Heathcote Ju 0.0 0.0 0.0 2.8 0.9
Warburton 0.0 0.0 0.0 1.8 7.6
Millgrove 0.0 0.0 6.5 2.5 5.4
Ben Cairn 0.0 0.0 0.0 2.5 7.4
Panton Gap 0.0 0.0 0.0 3.1 9.2
OShannassy 0.0 0.0 0.0 1.5 3.1
Ghin Ghin 8.4 8.4 3.4 0.0 0.0
Minto 15.4 0.0 0.0 0.0 3.3
Hawke 0.0 0.0 0.0 2.1 3.7
St Andrews 0.0 9.0 0.0 3.7 5.6
Nepean 0.0 0.0 3.7 0.0 4.0
Cape Schanck 0.0 0.0 6.4 0.0 4.6
Balnarring 0.0 0.0 9.1 0.0 3.9
Bittern 27.7 0.0 2.3 0.0 0.0
Bailieston 0.0 8.7 0.0 0.0 0.0
Donna Buang 0.0 0.0 0.0 1.5 7.4
Upper Yarra 0.0 0.0 0.0 1.3 2.3
Gembrook 0.0 0.0 3.6 0.0 26.6
Arcadia 27.6 0.0 4.3 3.7 0.0
Undera 13.5 11.5 2.0 0.0 0.0
Coomboona 13.9 5.6 4.6 1.1 0.0
Toolamba 16.0 0.0 5.0 0.0 0.0
Rushworth 0.0 10.7 3.2 0.0 0.0
Sayers 0.0 20.2 1.1 0.0 0.0
Waranga 25.3 2.2 3.4 0.0 0.0
Costerfield 0.0 15.8 1.1 0.0 0.0
Tallarook 0.0 2.9 0.0 0.0 5.8
SCR RBFT BFCS WAG WWCH
Reedy Lake 0.0 0.0 0.6 1.9 0.0
Pearcedale 0.0 1.1 1.1 3.4 0.0
Warneet 0.0 0.0 1.5 2.1 0.0
Cranbourne 0.0 5.0 1.4 3.4 0.0
Lysterfield 0.7 0.0 2.7 0.0 0.0
Red Hill 0.0 0.7 2.0 0.0 0.0
Devilbend 0.0 0.0 3.6 0.0 0.0
Olinda 0.0 0.7 0.0 0.0 0.0
Fern Tree Gum 0.0 1.9 0.6 0.0 0.0
Sherwin 3.0 0.0 1.2 0.0 9.8
Heathcote Ju 2.6 0.0 0.0 0.0 11.7
Warburton 0.0 0.9 1.5 0.0 0.0
Millgrove 2.0 5.4 2.2 0.0 0.0
Ben Cairn 0.0 0.0 0.0 0.0 0.0
Panton Gap 0.0 3.7 0.0 0.0 0.0
OShannassy 0.0 9.6 0.7 0.0 0.0
Ghin Ghin 0.0 44.7 0.4 1.3 0.0
Minto 0.0 10.5 0.0 0.0 0.0
Hawke 0.0 0.0 0.7 0.0 3.2
St Andrews 4.0 0.9 0.0 0.0 10.0
Nepean 2.0 0.0 1.1 0.0 0.0
Cape Schanck 0.0 3.4 0.0 0.0 0.0
Balnarring 0.0 2.7 1.0 0.0 0.0
Bittern 0.0 2.3 2.3 6.3 0.0
Bailieston 6.2 0.0 1.6 0.0 10.0
Donna Buang 0.0 0.0 1.5 0.0 0.0
Upper Yarra 0.0 6.4 0.9 0.0 0.0
Gembrook 4.7 2.8 0.0 0.0 0.0
Arcadia 0.0 0.6 2.1 4.9 8.0
Undera 0.0 0.0 1.9 2.5 6.5
Coomboona 0.0 0.0 6.9 3.3 5.6
Toolamba 0.0 0.8 3.0 3.5 5.0
Rushworth 1.1 2.7 1.1 0.0 9.6
Sayers 2.6 0.0 0.5 0.0 5.6
Waranga 0.0 10.9 1.6 2.4 8.9
Costerfield 5.5 0.0 1.3 0.0 5.7
Tallarook 5.6 0.0 1.5 0.0 2.8
NHHE VS CST BTR AMAG
Reedy Lake 0.0 0.0 1.7 12.5 8.6
Pearcedale 6.9 0.0 0.9 0.0 0.0
Warneet 3.0 0.0 1.5 0.0 0.0
Cranbourne 32.0 0.0 1.4 0.0 0.0
Lysterfield 6.4 0.0 0.0 0.0 0.0
Red Hill 2.2 5.4 0.0 0.0 0.0
Devilbend 5.6 5.6 4.6 0.0 0.0
Olinda 0.0 1.9 0.0 0.0 0.0
Fern Tree Gum 0.0 4.2 0.0 0.0 0.0
Sherwin 0.0 5.1 0.0 0.0 0.0
Heathcote Ju 0.0 0.0 0.0 0.0 0.0
Warburton 0.0 3.9 2.5 0.0 0.0
Millgrove 0.0 5.4 3.2 0.0 0.0
Ben Cairn 2.1 0.0 0.0 0.0 0.0
Panton Gap 0.0 0.0 0.0 0.0 0.0
OShannassy 0.0 0.0 0.0 0.0 0.0
Ghin Ghin 0.4 0.6 0.6 0.0 8.4
Minto 0.0 0.0 3.5 0.0 6.7
Hawke 0.0 0.0 1.5 0.0 0.0
St Andrews 0.0 0.0 2.7 0.0 0.0
Nepean 1.1 4.5 0.0 0.0 0.0
Cape Schanck 33.3 0.0 0.0 0.0 0.0
Balnarring 4.9 10.1 0.0 0.0 0.0
Bittern 2.6 0.0 1.3 0.0 0.0
Bailieston 0.0 2.5 0.0 0.0 0.0
Donna Buang 0.0 0.0 1.5 0.0 0.0
Upper Yarra 0.0 0.0 0.0 0.0 0.0
Gembrook 4.8 9.7 0.0 0.0 0.0
Arcadia 0.0 0.0 1.8 6.7 3.1
Undera 0.0 3.8 0.0 4.0 3.2
Coomboona 0.0 3.3 1.0 4.2 5.4
Toolamba 0.0 0.0 0.8 7.0 3.7
Rushworth 0.0 2.7 0.0 0.0 0.0
Sayers 0.0 0.0 0.0 0.0 0.0
Waranga 0.0 0.0 0.7 5.5 2.7
Costerfield 0.0 3.3 1.1 5.5 0.0
Tallarook 0.0 3.8 3.4 0.0 0.0
SCC RWH WSW STP YFHE
Reedy Lake 12.5 0.6 0.0 4.8 0.0
Pearcedale 0.0 2.3 5.7 0.0 1.1
Warneet 0.0 1.4 24.3 3.1 11.7
Cranbourne 0.0 0.0 10.0 4.0 0.0
Lysterfield 0.0 7.0 0.0 0.0 6.1
Red Hill 0.0 6.8 0.0 0.0 0.0
Devilbend 0.0 7.3 3.6 2.4 0.0
Olinda 0.0 9.1 0.0 0.0 0.0
Fern Tree Gum 0.0 4.5 0.0 0.0 0.0
Sherwin 0.0 6.3 0.0 3.5 2.3
Heathcote Ju 0.0 5.9 0.0 4.4 4.7
Warburton 0.0 0.0 0.0 2.7 6.2
Millgrove 0.0 8.8 0.0 2.2 5.4
Ben Cairn 0.0 0.0 0.0 0.0 0.9
Panton Gap 0.0 0.0 0.0 1.2 4.9
OShannassy 0.0 3.9 0.0 3.1 8.5
Ghin Ghin 47.6 6.1 4.7 1.2 6.7
Minto 80.5 5.0 0.0 5.0 26.7
Hawke 0.0 4.2 0.0 0.0 3.2
St Andrews 0.0 8.4 0.0 5.1 5.0
Nepean 0.0 3.3 0.0 0.0 1.0
Cape Schanck 0.0 2.6 0.0 0.0 3.4
Balnarring 0.0 4.9 0.0 0.0 1.9
Bittern 0.0 0.0 12.5 2.3 19.5
Bailieston 0.0 7.3 0.0 0.0 1.1
Donna Buang 0.0 0.0 0.0 2.2 0.0
Upper Yarra 0.0 7.0 0.0 6.4 3.9
Gembrook 0.0 10.9 0.0 0.0 20.2
Arcadia 24.0 0.0 2.7 8.2 0.0
Undera 16.0 1.5 1.0 8.7 0.0
Coomboona 30.4 1.1 0.0 8.1 0.0
Toolamba 29.9 0.0 6.0 4.5 0.0
Rushworth 0.0 4.3 0.0 1.1 14.4
Sayers 0.0 3.7 0.0 0.0 8.0
Waranga 0.0 1.4 3.4 2.7 16.3
Costerfield 0.0 6.2 0.0 6.6 0.6
Tallarook 0.0 9.5 0.0 1.9 5.8
WHIP GAL FHE BRTH SPP
Reedy Lake 0.0 4.8 26.2 0.0 0.0
Pearcedale 0.0 0.0 0.0 0.0 1.1
Warneet 0.0 0.0 0.0 0.0 4.6
Cranbourne 0.0 2.8 0.0 0.0 0.8
Lysterfield 0.0 0.0 0.0 0.0 5.4
Red Hill 0.0 0.0 0.0 0.0 3.4
Devilbend 0.0 0.0 0.0 0.0 0.0
Olinda 2.0 0.0 0.0 0.0 2.0
Fern Tree Gum 3.2 0.0 0.0 0.0 2.6
Sherwin 0.0 0.0 0.0 6.0 4.2
Heathcote Ju 0.0 0.0 0.0 0.0 3.7
Warburton 3.3 0.0 0.0 0.0 6.2
Millgrove 2.6 0.0 0.0 0.0 5.3
Ben Cairn 3.7 0.0 0.0 0.0 3.7
Panton Gap 3.8 0.0 0.0 0.0 1.9
OShannassy 2.9 0.0 0.0 0.0 2.2
Ghin Ghin 0.0 0.0 0.0 0.0 4.5
Minto 0.0 0.0 0.0 0.0 5.0
Hawke 0.0 0.0 0.0 5.2 3.7
St Andrews 0.0 0.0 0.0 10.0 5.1
Nepean 0.0 0.0 0.0 0.0 4.7
Cape Schanck 0.0 0.0 0.0 0.0 0.0
Balnarring 0.0 0.0 0.0 0.0 0.0
Bittern 0.0 0.0 0.0 0.0 3.5
Bailieston 0.0 0.0 1.2 0.0 2.8
Donna Buang 3.7 0.0 0.0 0.0 3.7
Upper Yarra 0.9 0.0 0.0 0.0 3.9
Gembrook 0.0 0.0 0.0 0.0 4.5
Arcadia 0.0 4.1 0.0 0.0 0.0
Undera 0.0 8.6 0.0 0.0 0.0
Coomboona 0.0 5.4 0.0 0.0 0.0
Toolamba 0.0 7.8 0.0 0.0 0.0
Rushworth 0.0 0.0 9.6 11.7 3.2
Sayers 0.0 0.0 3.1 9.1 5.7
Waranga 0.0 5.9 14.8 0.0 2.2
Costerfield 0.0 0.0 15.9 13.9 6.3
Tallarook 0.0 0.0 0.0 30.6 8.3
SIL GCU MUSK MGLK BHHE
Reedy Lake 0.0 0.0 13.1 1.7 1.1
Pearcedale 0.0 0.0 0.0 0.0 0.0
Warneet 0.0 0.0 0.0 0.0 0.0
Cranbourne 0.0 0.0 0.0 1.4 0.0
Lysterfield 0.0 0.0 0.0 0.0 0.0
Red Hill 2.7 1.4 0.0 0.0 0.0
Devilbend 1.2 0.0 0.0 0.0 0.0
Olinda 0.0 2.6 0.0 0.0 0.0
Fern Tree Gum 4.9 1.3 0.0 0.0 0.0
Sherwin 1.2 0.0 0.0 0.0 0.0
Heathcote Ju 2.5 1.5 0.0 0.0 0.0
Warburton 4.6 5.7 0.0 0.0 7.4
Millgrove 3.2 1.1 0.0 0.0 0.9
Ben Cairn 12.0 2.1 0.0 0.0 0.0
Panton Gap 2.4 2.9 0.0 0.0 7.7
OShannassy 3.7 0.0 0.0 0.0 0.0
Ghin Ghin 6.7 0.0 0.0 2.7 0.0
Minto 17.5 0.0 0.0 0.0 0.0
Hawke 0.0 0.0 0.0 0.0 0.5
St Andrews 0.9 3.4 0.0 0.0 1.0
Nepean 1.1 0.0 0.0 0.0 5.0
Cape Schanck 0.0 0.0 0.0 0.0 0.0
Balnarring 10.3 0.0 0.0 0.0 3.9
Bittern 8.0 0.0 0.0 0.0 2.3
Bailieston 0.8 0.0 0.0 0.0 14.1
Donna Buang 4.4 4.4 0.0 0.0 0.0
Upper Yarra 2.3 0.0 0.0 0.0 0.6
Gembrook 1.8 1.8 0.0 0.0 5.6
Arcadia 9.8 0.0 0.0 3.7 0.0
Undera 0.0 0.0 0.0 1.6 0.0
Coomboona 0.0 0.0 0.0 2.3 0.0
Toolamba 0.0 0.0 0.0 1.6 0.0
Rushworth 2.7 1.1 16.0 0.0 9.9
Sayers 3.7 1.6 3.1 0.0 7.8
Waranga 0.0 0.0 20.0 0.0 8.1
Costerfield 3.9 1.6 3.8 0.0 10.8
Tallarook 0.0 0.9 0.0 0.0 2.3
RFC YTBC LYRE CHE OWH
Reedy Lake 0.0 0.0 0.0 0.0 0.0
Pearcedale 0.0 0.0 0.0 0.0 0.0
Warneet 0.0 0.0 0.0 0.0 0.0
Cranbourne 0.0 0.0 0.0 0.0 0.0
Lysterfield 0.0 0.0 0.0 0.0 0.0
Red Hill 0.0 0.0 0.0 0.0 0.0
Devilbend 0.0 0.0 0.0 0.0 0.0
Olinda 0.0 1.9 0.0 0.0 0.0
Fern Tree Gum 0.0 2.6 0.6 0.6 0.0
Sherwin 0.0 0.0 0.0 0.0 0.0
Heathcote Ju 0.0 1.8 0.0 0.0 0.0
Warburton 0.0 3.9 1.4 0.0 0.0
Millgrove 0.0 1.9 0.0 2.2 0.0
Ben Cairn 0.0 3.7 0.0 5.6 3.7
Panton Gap 0.0 0.0 2.4 1.8 0.6
OShannassy 0.0 1.5 1.6 8.1 0.0
Ghin Ghin 1.3 0.0 0.0 0.0 0.0
Minto 2.8 0.0 0.0 0.0 2.8
Hawke 0.0 0.0 1.7 0.0 0.0
St Andrews 0.0 0.0 0.0 0.0 0.0
Nepean 0.0 0.0 0.0 5.6 0.0
Cape Schanck 0.0 0.0 0.0 5.2 0.0
Balnarring 0.0 0.0 0.0 0.0 0.0
Bittern 0.0 0.0 0.0 0.0 0.0
Bailieston 0.0 0.0 0.0 0.0 0.0
Donna Buang 0.0 3.6 3.0 1.5 0.7
Upper Yarra 0.0 0.8 0.9 0.9 0.0
Gembrook 0.0 5.5 0.0 11.2 0.0
Arcadia 4.3 0.0 0.0 0.0 0.0
Undera 1.5 0.0 0.0 0.0 0.0
Coomboona 2.6 0.0 0.0 0.0 0.0
Toolamba 2.0 0.0 0.0 0.0 0.0
Rushworth 0.0 0.0 0.0 0.0 0.0
Sayers 0.0 0.0 0.0 0.0 0.0
Waranga 2.7 0.0 0.0 0.0 0.0
Costerfield 0.0 0.0 0.0 0.0 0.0
Tallarook 0.0 0.0 0.0 0.0 0.0
TRM MB STHR LHE FTC
Reedy Lake 15.0 0.0 0.0 0.0 0.0
Pearcedale 0.0 0.0 0.0 0.0 2.3
Warneet 0.0 0.0 0.0 0.0 0.0
Cranbourne 0.0 1.0 0.0 0.0 0.0
Lysterfield 0.0 0.0 0.0 0.0 2.1
Red Hill 0.0 0.0 0.0 0.0 0.0
Devilbend 0.0 3.6 0.0 0.0 0.0
Olinda 0.0 0.0 0.0 0.0 2.6
Fern Tree Gum 0.0 0.0 0.0 0.0 2.6
Sherwin 0.0 1.2 0.0 0.0 1.7
Heathcote Ju 0.0 0.0 0.9 0.0 1.6
Warburton 0.0 0.0 1.4 2.1 2.1
Millgrove 0.0 0.0 0.0 0.0 3.5
Ben Cairn 0.0 0.0 0.0 4.1 4.6
Panton Gap 0.0 0.0 0.9 1.8 3.1
OShannassy 0.0 0.0 0.0 2.4 5.4
Ghin Ghin 0.0 0.0 0.0 0.0 2.4
Minto 0.0 0.0 0.0 0.0 1.7
Hawke 0.0 0.0 0.0 0.0 1.1
St Andrews 0.0 1.7 0.0 0.0 3.4
Nepean 0.0 2.2 0.0 0.0 1.9
Cape Schanck 0.0 0.0 0.0 0.0 1.7
Balnarring 0.0 4.9 0.0 0.0 1.0
Bittern 0.0 0.0 0.0 0.0 2.3
Bailieston 0.0 0.0 0.0 0.0 0.0
Donna Buang 0.0 0.0 0.0 2.2 2.2
Upper Yarra 0.0 0.0 0.0 0.9 2.3
Gembrook 0.0 0.8 2.4 1.9 2.8
Arcadia 2.5 0.0 0.0 0.0 0.6
Undera 0.0 0.5 0.0 0.0 0.0
Coomboona 0.6 0.0 0.0 0.0 0.0
Toolamba 3.3 0.0 0.0 0.0 0.0
Rushworth 0.0 0.0 0.0 0.0 0.0
Sayers 0.0 0.0 0.0 0.0 0.0
Waranga 4.8 0.0 0.0 0.0 0.0
Costerfield 0.0 0.0 0.0 0.0 1.6
Tallarook 0.0 0.0 0.0 0.0 2.9
PINK OBO YR LFB SPW RBTR
Reedy Lake 0.0 0.0 0.0 2.9 0.0 0.0
Pearcedale 0.0 0.0 0.0 0.0 0.0 0.0
Warneet 0.0 0.0 0.0 0.0 0.0 0.0
Cranbourne 0.0 0.0 0.0 0.0 0.0 0.0
Lysterfield 0.0 1.4 0.0 0.0 0.0 0.0
Red Hill 0.0 0.0 0.0 0.0 0.0 0.0
Devilbend 0.0 0.0 0.0 0.0 0.0 0.0
Olinda 0.0 0.0 0.0 0.0 0.0 0.0
Fern Tree Gum 0.0 0.0 0.0 0.0 0.0 0.0
Sherwin 0.0 1.2 0.0 0.0 0.0 0.6
Heathcote Ju 0.0 0.0 0.0 0.0 0.0 1.6
Warburton 0.0 0.0 0.0 0.0 0.0 0.0
Millgrove 0.0 0.0 0.0 0.0 0.0 1.1
Ben Cairn 0.0 0.0 0.0 0.0 0.0 0.0
Panton Gap 1.0 0.0 0.0 0.0 0.0 0.0
OShannassy 2.2 0.0 0.0 0.0 0.0 0.0
Ghin Ghin 0.0 1.2 0.0 0.0 0.0 0.0
Minto 0.0 0.0 0.0 0.0 0.0 0.0
Hawke 0.0 0.0 0.0 0.0 0.0 1.7
St Andrews 0.0 0.0 0.0 0.0 0.0 0.0
Nepean 0.0 0.0 0.0 0.0 0.0 0.0
Cape Schanck 0.0 0.0 0.0 0.0 0.0 0.0
Balnarring 0.0 0.0 0.0 0.0 0.0 0.0
Bittern 0.0 0.0 0.0 0.0 0.0 0.0
Bailieston 0.0 3.3 0.0 0.0 0.0 0.0
Donna Buang 0.8 0.0 0.0 0.0 0.0 0.0
Upper Yarra 0.0 0.0 0.0 0.0 0.0 0.0
Gembrook 0.0 0.0 0.0 0.0 0.0 0.0
Arcadia 0.0 2.5 0.0 1.4 0.0 0.0
Undera 0.0 0.0 3.2 1.0 0.0 0.0
Coomboona 0.0 0.0 2.6 5.9 0.0 0.0
Toolamba 0.0 2.0 0.0 6.6 0.0 0.0
Rushworth 0.0 1.1 0.0 0.0 1.1 0.0
Sayers 0.0 0.5 0.0 0.0 0.0 0.0
Waranga 0.0 2.4 0.0 0.0 0.0 0.0
Costerfield 0.0 1.1 0.0 0.0 3.3 0.0
Tallarook 0.0 0.0 0.0 0.0 2.9 0.0
DWS BELL LWB CBW GGC PIL
Reedy Lake 0.4 0.0 0.0 0.0 0.0 0.0
Pearcedale 0.0 0.0 0.0 0.0 0.0 0.0
Warneet 3.5 0.0 0.0 0.0 0.0 0.0
Cranbourne 5.5 0.0 4.0 0.0 0.0 0.0
Lysterfield 0.0 22.1 0.0 0.0 0.0 0.0
Red Hill 0.0 0.0 0.0 0.0 0.0 0.0
Devilbend 1.8 0.0 0.0 0.0 0.0 0.0
Olinda 0.0 0.0 0.0 0.0 0.0 0.0
Fern Tree Gum 0.0 0.0 0.0 0.0 1.3 1.3
Sherwin 0.0 0.0 0.0 0.6 0.0 0.0
Heathcote Ju 0.0 0.0 0.0 0.0 0.0 0.0
Warburton 0.0 0.0 0.0 0.0 1.8 0.7
Millgrove 0.9 0.0 0.0 0.0 0.0 0.0
Ben Cairn 0.0 0.0 0.0 0.0 3.7 2.8
Panton Gap 0.0 0.0 0.0 0.0 4.8 1.8
OShannassy 0.0 0.0 0.0 0.0 0.0 2.2
Ghin Ghin 0.0 0.0 0.0 0.0 0.0 0.0
Minto 0.0 0.0 0.0 0.0 0.0 0.0
Hawke 0.0 0.0 0.0 0.0 0.0 0.0
St Andrews 0.9 15.0 0.0 0.0 0.0 0.0
Nepean 0.0 0.0 0.0 0.0 0.0 0.0
Cape Schanck 0.0 0.0 32.2 0.0 0.0 0.0
Balnarring 0.0 0.0 16.5 1.0 0.0 0.0
Bittern 0.0 0.0 5.8 0.0 0.0 0.0
Bailieston 0.0 0.0 0.0 1.7 0.0 0.0
Donna Buang 0.0 0.0 0.0 0.0 0.7 4.4
Upper Yarra 0.7 0.0 0.0 0.0 0.0 0.0
Gembrook 0.0 0.0 0.0 0.0 0.0 0.0
Arcadia 0.0 0.0 0.0 0.0 0.0 0.0
Undera 0.0 0.0 0.0 0.0 0.0 0.0
Coomboona 0.0 0.0 0.0 0.0 0.0 0.0
Toolamba 0.0 0.0 0.0 0.0 0.0 0.0
Rushworth 0.0 0.0 0.0 0.0 0.0 0.0
Sayers 0.0 0.0 0.0 1.6 0.0 0.0
Waranga 4.8 0.0 0.0 0.0 0.0 0.0
Costerfield 0.6 0.0 0.0 0.0 0.0 0.0
Tallarook 0.0 0.0 0.0 0.0 0.0 0.0
SKF RSL PDOV CRP JW
Reedy Lake 1.9 6.7 0.0 0.0 0.0
Pearcedale 0.0 0.0 0.0 0.0 0.0
Warneet 0.0 0.0 0.0 0.0 0.0
Cranbourne 0.0 0.8 0.0 0.0 0.0
Lysterfield 0.0 0.0 0.0 0.0 0.0
Red Hill 0.0 0.0 0.0 0.0 0.0
Devilbend 0.0 0.0 0.0 0.0 0.0
Olinda 0.0 0.0 0.0 0.0 0.0
Fern Tree Gum 0.0 0.0 0.0 0.0 0.0
Sherwin 2.3 0.0 0.0 0.0 0.0
Heathcote Ju 0.0 0.0 0.0 0.0 0.0
Warburton 0.0 0.0 0.0 0.0 0.0
Millgrove 0.0 0.0 0.0 0.0 0.0
Ben Cairn 0.0 0.0 0.0 0.0 0.0
Panton Gap 0.0 0.0 0.0 0.0 0.0
OShannassy 0.0 0.0 0.0 0.0 0.0
Ghin Ghin 1.8 0.0 0.0 0.0 0.0
Minto 1.7 0.0 0.0 0.0 0.0
Hawke 0.0 0.0 0.0 0.0 0.0
St Andrews 2.0 0.0 0.0 0.0 0.0
Nepean 0.0 0.0 0.0 0.0 0.0
Cape Schanck 0.0 0.0 0.0 0.0 0.0
Balnarring 0.0 0.0 0.0 0.0 0.0
Bittern 0.0 0.0 0.0 0.0 0.0
Bailieston 0.0 0.0 0.0 0.0 0.0
Donna Buang 0.0 0.0 0.0 0.0 0.0
Upper Yarra 1.6 0.0 0.0 0.0 0.0
Gembrook 0.0 0.0 0.0 0.0 0.0
Arcadia 2.1 11.0 3.1 1.8 1.2
Undera 2.1 0.0 0.0 0.0 3.8
Coomboona 2.2 1.1 0.0 1.1 2.8
Toolamba 1.0 5.0 0.4 0.0 0.0
Rushworth 0.0 1.4 0.0 0.0 0.0
Sayers 0.0 1.7 0.0 0.0 0.0
Waranga 1.4 0.0 0.8 0.0 0.0
Costerfield 0.0 0.0 0.0 0.0 0.0
Tallarook 1.7 0.0 0.0 0.0 0.0
BCHE RCR GBB RRP LLOR
Reedy Lake 0.0 0.0 0.0 4.8 0.0
Pearcedale 0.0 0.0 0.0 0.0 0.0
Warneet 0.0 0.0 0.0 0.0 0.0
Cranbourne 0.0 0.0 0.0 0.0 0.0
Lysterfield 0.0 0.0 0.7 0.0 0.0
Red Hill 0.0 0.0 0.0 0.0 0.0
Devilbend 0.0 0.0 0.0 0.0 0.0
Olinda 0.0 0.0 0.0 0.0 0.0
Fern Tree Gum 0.0 0.0 0.6 0.0 0.0
Sherwin 0.0 0.0 0.0 0.0 0.0
Heathcote Ju 0.0 0.0 0.0 0.0 0.0
Warburton 0.0 0.0 0.0 0.0 0.0
Millgrove 0.0 0.0 0.0 0.0 0.0
Ben Cairn 0.0 0.0 0.0 0.0 0.0
Panton Gap 0.0 0.0 0.0 0.0 0.0
OShannassy 0.0 0.0 0.0 0.0 0.0
Ghin Ghin 0.0 0.0 0.0 0.0 0.0
Minto 0.0 0.0 0.0 0.0 0.0
Hawke 0.0 0.0 0.0 0.0 0.0
St Andrews 0.0 0.0 0.0 0.0 0.0
Nepean 0.0 0.0 0.0 0.0 0.0
Cape Schanck 0.0 0.0 0.0 0.0 0.0
Balnarring 0.0 0.0 0.0 0.0 0.0
Bittern 0.0 0.0 0.0 0.0 0.0
Bailieston 0.0 3.3 0.0 0.0 0.0
Donna Buang 0.0 0.0 0.0 0.0 0.0
Upper Yarra 0.0 0.0 0.9 0.0 0.0
Gembrook 0.0 0.0 0.0 0.0 0.0
Arcadia 0.0 0.0 0.0 0.0 0.0
Undera 0.0 0.0 0.0 0.0 0.0
Coomboona 0.0 0.0 0.0 0.0 0.0
Toolamba 1.2 0.0 0.0 0.0 0.0
Rushworth 0.0 0.9 0.0 0.0 0.0
Sayers 0.0 0.5 0.5 0.0 0.0
Waranga 0.0 0.0 0.8 4.8 10.9
Costerfield 0.0 0.5 0.0 0.0 0.0
Tallarook 0.0 0.0 0.0 0.0 0.0
YTHE RF SHBC AZKF SFC
Reedy Lake 0.0 0.0 0.0 0.0 0.0
Pearcedale 0.0 0.0 0.0 0.0 0.0
Warneet 0.0 0.0 1.4 0.0 0.0
Cranbourne 0.0 0.0 0.0 0.0 0.0
Lysterfield 0.0 0.0 0.7 0.0 0.0
Red Hill 0.0 1.4 0.0 0.0 3.4
Devilbend 0.0 0.0 0.0 0.0 0.0
Olinda 0.0 1.9 2.0 0.7 1.2
Fern Tree Gum 0.0 3.2 1.3 0.0 1.9
Sherwin 0.0 0.0 0.6 0.0 1.2
Heathcote Ju 0.0 0.0 1.5 0.0 1.6
Warburton 0.0 1.8 2.1 0.0 4.1
Millgrove 0.0 1.1 0.0 0.0 7.6
Ben Cairn 0.0 4.6 1.9 0.0 2.8
Panton Gap 0.0 1.8 3.7 0.0 1.8
OShannassy 0.0 1.6 1.6 0.0 0.0
Ghin Ghin 0.0 0.0 1.8 0.0 0.0
Minto 0.0 0.0 0.0 0.0 0.0
Hawke 0.0 0.0 0.0 0.0 0.0
St Andrews 0.0 0.0 0.0 0.0 0.0
Nepean 0.0 2.2 1.9 0.0 0.0
Cape Schanck 0.0 2.6 0.9 0.0 0.0
Balnarring 0.0 0.0 1.9 0.0 0.0
Bittern 0.0 0.0 0.0 0.0 0.0
Bailieston 0.0 0.0 0.0 0.0 0.0
Donna Buang 0.0 2.2 0.7 0.0 0.7
Upper Yarra 0.0 0.0 1.6 0.0 0.8
Gembrook 0.0 2.8 1.8 0.0 0.0
Arcadia 0.0 0.0 0.0 0.0 0.0
Undera 0.0 0.0 0.0 0.0 0.0
Coomboona 0.0 0.0 0.0 0.0 0.0
Toolamba 0.0 0.0 0.0 0.0 0.0
Rushworth 0.0 0.0 0.0 0.0 0.0
Sayers 0.0 0.0 0.0 0.0 0.0
Waranga 2.7 0.0 0.0 0.0 0.0
Costerfield 6.3 0.0 1.6 0.0 0.0
Tallarook 0.0 0.0 1.9 0.0 0.0
YRTH ROSE BCOO LFC WG
Reedy Lake 0.0 0.0 0.0 0.0 0.0
Pearcedale 0.0 0.0 0.0 0.0 0.0
Warneet 0.0 0.0 0.0 1.8 0.0
Cranbourne 0.0 0.0 0.0 0.0 0.0
Lysterfield 0.0 0.0 0.0 0.0 0.0
Red Hill 0.0 0.0 0.0 0.0 0.0
Devilbend 0.0 0.0 0.0 2.4 0.0
Olinda 0.0 0.6 0.0 0.0 0.0
Fern Tree Gum 0.0 0.0 0.6 0.0 0.0
Sherwin 0.0 0.0 0.0 0.0 0.0
Heathcote Ju 0.0 0.0 0.0 0.0 0.0
Warburton 0.0 0.0 0.0 0.0 0.0
Millgrove 0.0 0.0 0.0 0.0 0.0
Ben Cairn 0.0 1.9 2.8 0.0 0.0
Panton Gap 0.0 3.1 1.2 0.0 0.0
OShannassy 0.0 0.7 1.6 0.0 0.0
Ghin Ghin 3.9 0.0 0.0 1.2 0.0
Minto 0.0 0.0 0.0 0.0 0.0
Hawke 0.0 0.0 0.0 0.0 0.0
St Andrews 0.0 0.0 0.0 0.0 0.0
Nepean 0.0 0.0 0.0 0.0 0.0
Cape Schanck 0.0 0.0 0.0 0.0 0.0
Balnarring 0.0 0.0 0.0 0.0 0.0
Bittern 0.0 0.0 0.0 0.0 0.0
Bailieston 0.0 0.0 0.0 0.0 0.0
Donna Buang 0.0 1.5 0.0 0.0 0.0
Upper Yarra 0.0 0.0 0.0 0.0 0.0
Gembrook 0.0 0.0 0.0 5.5 0.0
Arcadia 0.0 0.0 0.0 2.5 0.0
Undera 5.9 0.0 0.0 0.0 3.1
Coomboona 0.0 0.0 0.0 0.0 1.6
Toolamba 0.0 0.0 0.0 0.0 0.0
Rushworth 0.0 0.0 0.0 0.0 0.5
Sayers 0.0 0.0 0.0 0.0 0.0
Waranga 0.0 0.0 0.0 0.0 0.0
Costerfield 0.0 0.0 0.0 0.0 0.0
Tallarook 0.0 0.0 0.0 0.0 0.0
PCOO WTG NMIN NFB DB
Reedy Lake 1.9 0.0 0.2 0.0 0.0
Pearcedale 0.0 0.0 0.0 0.0 0.0
Warneet 0.0 0.0 5.8 0.0 0.0
Cranbourne 0.0 0.0 3.1 0.0 0.0
Lysterfield 0.0 0.0 0.0 0.0 0.0
Red Hill 0.0 0.0 0.0 0.0 0.0
Devilbend 0.0 0.0 0.0 0.0 0.0
Olinda 0.0 0.0 0.0 0.0 0.0
Fern Tree Gum 0.0 0.0 0.0 0.0 0.0
Sherwin 0.0 0.0 0.0 0.0 0.0
Heathcote Ju 0.0 0.0 0.0 0.0 0.0
Warburton 0.0 0.0 0.0 0.0 0.0
Millgrove 0.0 0.0 0.0 0.0 0.0
Ben Cairn 0.0 0.0 0.0 0.0 0.0
Panton Gap 0.0 0.0 0.0 0.0 0.0
OShannassy 0.0 0.0 0.0 0.0 0.0
Ghin Ghin 0.0 0.8 0.0 1.8 1.2
Minto 0.0 0.0 0.0 0.0 0.0
Hawke 0.0 0.0 0.0 0.0 0.0
St Andrews 0.0 0.0 0.0 0.0 0.0
Nepean 0.0 0.0 0.0 0.0 0.0
Cape Schanck 0.0 0.0 0.0 0.0 0.0
Balnarring 0.0 0.0 0.0 0.0 0.0
Bittern 0.0 0.0 0.0 0.0 0.0
Bailieston 0.0 3.1 0.0 0.0 0.0
Donna Buang 0.0 0.0 0.0 0.0 0.0
Upper Yarra 0.0 0.0 0.0 0.0 0.0
Gembrook 0.0 0.0 0.0 0.0 0.0
Arcadia 0.0 0.0 0.0 0.0 1.4
Undera 0.0 3.1 0.0 1.5 0.0
Coomboona 0.0 1.6 5.4 0.0 1.6
Toolamba 0.0 0.0 5.7 0.8 1.5
Rushworth 0.0 0.5 0.0 0.0 0.0
Sayers 0.0 0.0 0.0 0.0 0.0
Waranga 0.0 0.0 1.4 2.4 0.0
Costerfield 0.0 0.0 0.0 0.0 0.0
Tallarook 0.0 0.0 0.0 0.0 0.0
RBEE HBC DF PCL FLAME
Reedy Lake 0.0 0.0 0.0 9.1 0.0
Pearcedale 0.0 0.0 0.0 0.0 0.0
Warneet 0.0 0.0 0.0 0.0 0.0
Cranbourne 0.0 0.0 0.0 0.0 0.0
Lysterfield 0.0 0.0 0.0 0.0 0.0
Red Hill 0.0 0.0 0.0 0.0 0.0
Devilbend 0.0 0.0 0.0 0.0 0.0
Olinda 0.0 0.0 0.0 0.0 0.0
Fern Tree Gum 0.0 0.0 0.0 0.0 0.0
Sherwin 0.0 0.0 0.0 0.0 0.0
Heathcote Ju 0.0 0.0 0.0 0.0 0.0
Warburton 0.0 0.0 0.0 0.0 0.0
Millgrove 0.0 0.0 0.0 0.0 0.0
Ben Cairn 0.0 0.0 0.0 0.0 0.0
Panton Gap 0.0 0.0 0.0 0.0 0.0
OShannassy 0.0 0.0 0.0 0.0 0.0
Ghin Ghin 0.0 0.0 0.0 0.0 2.6
Minto 0.0 0.0 0.0 0.0 0.0
Hawke 0.0 0.0 0.0 0.0 0.0
St Andrews 0.0 0.0 0.0 0.0 0.0
Nepean 0.0 0.0 0.0 0.0 0.0
Cape Schanck 0.0 0.0 0.0 0.0 0.0
Balnarring 0.0 0.0 0.0 0.0 1.9
Bittern 0.0 0.0 0.0 0.0 0.0
Bailieston 0.0 0.0 0.0 0.0 0.0
Donna Buang 0.0 0.0 0.0 0.0 2.9
Upper Yarra 0.0 0.0 0.0 0.0 0.0
Gembrook 0.0 0.0 0.0 0.0 0.0
Arcadia 1.4 0.0 0.0 0.0 1.8
Undera 1.0 0.0 0.0 0.0 5.9
Coomboona 0.0 0.0 0.0 0.0 1.7
Toolamba 0.5 0.4 0.0 0.0 0.0
Rushworth 0.0 0.5 0.0 0.0 0.0
Sayers 0.0 0.0 0.0 0.0 0.0
Waranga 0.7 0.0 2.1 0.0 0.0
Costerfield 1.1 0.0 1.1 0.0 0.0
Tallarook 0.0 0.0 3.8 0.0 0.0
WWT WBWS LCOR KING
Reedy Lake 0.0 0.0 0.0 0.0
Pearcedale 0.0 0.0 0.0 0.0
Warneet 0.0 0.0 0.0 0.0
Cranbourne 0.0 0.0 0.0 0.0
Lysterfield 0.0 0.0 0.0 0.0
Red Hill 0.0 0.0 0.0 0.0
Devilbend 0.0 0.0 0.0 0.0
Olinda 0.0 0.0 0.0 0.0
Fern Tree Gum 0.0 0.0 0.0 0.0
Sherwin 0.0 0.0 0.0 0.0
Heathcote Ju 0.0 0.0 0.0 0.0
Warburton 0.0 0.0 0.0 0.8
Millgrove 0.0 0.0 0.0 0.0
Ben Cairn 0.0 0.0 0.0 0.0
Panton Gap 0.0 0.0 0.0 0.0
OShannassy 0.0 0.0 0.0 0.7
Ghin Ghin 0.0 0.0 0.0 0.0
Minto 0.0 0.0 0.0 0.0
Hawke 0.0 0.0 0.0 0.0
St Andrews 0.0 0.0 0.0 0.0
Nepean 0.0 0.0 0.0 0.0
Cape Schanck 0.0 0.0 0.0 0.0
Balnarring 0.0 0.0 0.0 0.0
Bittern 0.0 0.0 0.0 0.0
Bailieston 0.0 0.0 0.0 0.0
Donna Buang 0.0 0.0 0.0 0.0
Upper Yarra 0.0 0.0 0.0 0.0
Gembrook 0.0 0.0 0.0 0.0
Arcadia 2.1 0.0 4.8 0.0
Undera 0.0 0.0 0.0 0.0
Coomboona 0.0 0.0 0.0 0.0
Toolamba 2.5 0.0 0.0 0.0
Rushworth 0.0 0.0 0.0 0.0
Sayers 0.0 0.0 0.0 0.0
Waranga 0.0 1.6 0.0 0.0
Costerfield 0.0 0.0 0.0 0.0
Tallarook 0.0 0.0 0.0 0.0
- It is highly likely that some species will be more abundant than others - some will be very common and others will be rare.
Furthermore, some sites may have similar compositions yet vastly different total abundances. There might be numereous ecological
explanations for such situations, yet we are primarily interested in describing the community compositions.
- Calculate the means and maximums of each species
Show code
> apply(macnally[,c(-1,-2)],2,mean, na.rm=TRUE)
EYR GF BTH GWH 3.71081 6.11622 14.70270 3.63784 WTTR WEHE WNHE SFW 4.50811 3.56216 9.14865 5.98919 WBSW CR LK RWB 5.06757 5.38919 2.90000 5.67297 AUR STTH LR WPHE 2.17297 7.98378 3.41622 7.21351 YTH ER PCU ESP 2.82703 2.25135 0.90000 3.55946 SCR RBFT BFCS WAG 1.08108 3.18108 1.31081 0.94595 WWCH NHHE VS CST 2.76757 2.84595 2.04865 0.96486 BTR AMAG SCC RWH 1.22703 1.12973 6.51081 4.08919 WSW STP YFHE WHIP 1.99730 2.57838 5.12162 0.70541 GAL FHE BRTH SPP 1.06486 1.91351 2.33784 3.08649 SIL GCU MUSK MGLK 3.03514 0.90270 1.51351 0.40541 BHHE RFC YTBC LYRE 2.40541 0.46486 0.73514 0.31351 CHE OWH TRM MB 1.15405 0.21081 0.70811 0.42973 STHR LHE FTC PINK 0.15135 0.41622 1.60811 0.10811 OBO YR LFB SPW 0.45135 0.15676 0.48108 0.19730 RBTR DWS BELL LWB 0.13514 0.51622 1.00270 1.58108 CBW GGC PIL SKF 0.13243 0.33243 0.35676 0.58919 RSL PDOV CRP JW 0.74865 0.11622 0.07838 0.21081 BCHE RCR GBB RRP 0.03243 0.14054 0.09459 0.25946 LLOR YTHE RF SHBC 0.29459 0.24324 0.73514 0.83514 AZKF SFC YRTH ROSE 0.01892 0.73243 0.26486 0.21081 BCOO LFC WG PCOO 0.16757 0.36216 0.14054 0.05135 WTG NMIN NFB DB 0.24595 0.58378 0.17568 0.15405 RBEE HBC DF PCL 0.12703 0.02432 0.18919 0.24595 FLAME WWT WBWS LCOR 0.45405 0.12432 0.04324 0.12973 KING 0.04054> apply(macnally[,c(-1,-2)],2,max)
EYR GF BTH GWH WTTR WEHE 9.7 13.3 36.1 9.5 12.8 11.5 WNHE SFW WBSW CR LK RWB 38.9 22.0 12.6 19.1 6.7 26.3 AUR STTH LR WPHE YTH ER 8.8 25.2 16.5 27.7 20.2 9.1 PCU ESP SCR RBFT BFCS WAG 3.7 26.6 6.2 44.7 6.9 6.3 WWCH NHHE VS CST BTR AMAG 11.7 33.3 10.1 4.6 12.5 8.6 SCC RWH WSW STP YFHE WHIP 80.5 10.9 24.3 8.7 26.7 3.8 GAL FHE BRTH SPP SIL GCU 8.6 26.2 30.6 8.3 17.5 5.7 MUSK MGLK BHHE RFC YTBC LYRE 20.0 3.7 14.1 4.3 5.5 3.0 CHE OWH TRM MB STHR LHE 11.2 3.7 15.0 4.9 2.4 4.1 FTC PINK OBO YR LFB SPW 5.4 2.2 3.3 3.2 6.6 3.3 RBTR DWS BELL LWB CBW GGC 1.7 5.5 22.1 32.2 1.7 4.8 PIL SKF RSL PDOV CRP JW 4.4 2.3 11.0 3.1 1.8 3.8 BCHE RCR GBB RRP LLOR YTHE 1.2 3.3 0.9 4.8 10.9 6.3 RF SHBC AZKF SFC YRTH ROSE 4.6 3.7 0.7 7.6 5.9 3.1 BCOO LFC WG PCOO WTG NMIN 2.8 5.5 3.1 1.9 3.1 5.8 NFB DB RBEE HBC DF PCL 2.4 1.6 1.4 0.5 3.8 9.1 FLAME WWT WBWS LCOR KING 5.9 2.5 1.6 4.8 0.8
> apply(macnally[,c(-1,-2)],2,sum)
EYR GF BTH GWH WTTR WEHE 137.3 226.3 544.0 134.6 166.8 131.8 WNHE SFW WBSW CR LK RWB 338.5 221.6 187.5 199.4 107.3 209.9 AUR STTH LR WPHE YTH ER 80.4 295.4 126.4 266.9 104.6 83.3 PCU ESP SCR RBFT BFCS WAG 33.3 131.7 40.0 117.7 48.5 35.0 WWCH NHHE VS CST BTR AMAG 102.4 105.3 75.8 35.7 45.4 41.8 SCC RWH WSW STP YFHE WHIP 240.9 151.3 73.9 95.4 189.5 26.1 GAL FHE BRTH SPP SIL GCU 39.4 70.8 86.5 114.2 112.3 33.4 MUSK MGLK BHHE RFC YTBC LYRE 56.0 15.0 89.0 17.2 27.2 11.6 CHE OWH TRM MB STHR LHE 42.7 7.8 26.2 15.9 5.6 15.4 FTC PINK OBO YR LFB SPW 59.5 4.0 16.7 5.8 17.8 7.3 RBTR DWS BELL LWB CBW GGC 5.0 19.1 37.1 58.5 4.9 12.3 PIL SKF RSL PDOV CRP JW 13.2 21.8 27.7 4.3 2.9 7.8 BCHE RCR GBB RRP LLOR YTHE 1.2 5.2 3.5 9.6 10.9 9.0 RF SHBC AZKF SFC YRTH ROSE 27.2 30.9 0.7 27.1 9.8 7.8 BCOO LFC WG PCOO WTG NMIN 6.2 13.4 5.2 1.9 9.1 21.6 NFB DB RBEE HBC DF PCL 6.5 5.7 4.7 0.9 7.0 9.1 FLAME WWT WBWS LCOR KING 16.8 4.6 1.6 4.8 1.5
> apply(macnally[,c(-1,-2)],2,var, na.rm=TRUE)
EYR GF BTH GWH 7.71988 17.85306 124.66083 7.38686 WTTR WEHE WNHE SFW 10.16465 10.08520 74.11923 25.76432 WBSW CR LK RWB 17.33892 29.98488 2.40556 53.48480 AUR STTH LR WPHE 3.45592 42.44806 16.91695 114.58953 YTH ER PCU ESP 27.02036 5.44090 1.62944 22.94581 SCR RBFT BFCS WAG 3.53102 58.37213 1.72766 2.73811 WWCH NHHE VS CST 15.70670 56.29366 7.71590 1.53734 BTR AMAG SCC RWH 8.01592 5.89381 277.33266 10.75321 WSW STP YFHE WHIP 22.84805 6.97841 43.93785 1.79219 GAL FHE BRTH SPP 5.68068 31.27842 36.39131 4.84565 SIL GCU MUSK MGLK 16.06790 1.95360 21.30287 0.83775 BHHE RFC YTBC LYRE 14.60164 1.12012 2.00623 0.54731 CHE OWH TRM MB 6.75089 0.57599 6.86077 1.14437 STHR LHE FTC PINK 0.23590 0.91862 2.03021 0.16799 OBO YR LFB SPW 0.74646 0.44697 2.25491 0.52971 RBTR DWS BELL LWB 0.17623 1.70973 18.78360 35.17324 CBW GGC PIL SKF 0.17114 1.05725 0.90974 0.78488 RSL PDOV CRP JW 4.91257 0.27529 0.11730 0.61321 BCHE RCR GBB RRP 0.03892 0.31859 0.06164 1.21081 LLOR YTHE RF SHBC 3.21108 1.24419 1.43068 0.91512 AZKF SFC YRTH ROSE 0.01324 2.43170 1.31734 0.40766 BCOO LFC WG PCOO 0.31003 1.16908 0.32470 0.09757 WTG NMIN NFB DB 0.56755 2.61917 0.29856 0.20366 RBEE HBC DF PCL 0.11980 0.01078 0.52044 2.23811 FLAME WWT WBWS LCOR 1.44700 0.28023 0.06919 0.62270 KING 0.02970 - Perform this standardization on the bird abundance data and confirm that this has evened out the abundances
Show code
> library(vegan) > macnally.std <- wisconsin(macnally[,c(-1,-2)]) > apply(macnally.std[,c(-1,-2)],2,max)
BTH GWH WTTR WEHE WNHE 0.06781 0.07385 0.08452 0.07973 0.05925 SFW WBSW CR LK RWB 0.07502 0.14357 0.07300 0.09260 0.08625 AUR STTH LR WPHE YTH 0.07300 0.06173 0.08024 0.14305 0.10025 ER PCU ESP SCR RBFT 0.08024 0.06672 0.04966 0.07914 0.05685 BFCS WAG WWCH NHHE VS 0.05846 0.07973 0.08486 0.09702 0.08024 CST BTR AMAG SCC RWH 0.08412 0.06491 0.06491 0.05302 0.07101 WSW STP YFHE WHIP GAL 0.08459 0.06218 0.05823 0.07076 0.05855 FHE BRTH SPP SIL GCU 0.06491 0.06817 0.06885 0.05302 0.05610 MUSK MGLK BHHE RFC YTBC 0.06900 0.04788 0.07914 0.04788 0.04966 LYRE CHE OWH TRM MB 0.07267 0.04966 0.04963 0.06491 0.08024 STHR LHE FTC PINK OBO 0.04966 0.04963 0.06217 0.06217 0.07914 YR LFB SPW RBTR DWS 0.05855 0.06309 0.06011 0.10713 0.09126 BELL LWB CBW GGC PIL 0.07300 0.09702 0.09435 0.05566 0.07267 SKF RSL PDOV CRP JW 0.08512 0.04788 0.04788 0.04788 0.05855 BCHE RCR GBB RRP LLOR 0.06309 0.07914 0.08452 0.06491 0.04603 YTHE RF SHBC AZKF SFC 0.06011 0.05484 0.05566 0.08505 0.06123 YRTH ROSE BCOO LFC WG 0.05855 0.05566 0.04963 0.04966 0.05855 PCOO WTG NMIN NFB DB 0.06491 0.07914 0.08459 0.04603 0.05915 RBEE HBC DF PCL FLAME 0.04788 0.08625 0.06817 0.06491 0.05855 WWT WBWS LCOR KING 0.06309 0.04603 0.04788 0.05440> apply(macnally.std[,c(-1,-2)],2,var, na.rm=TRUE)
BTH GWH WTTR WEHE 4.704e-04 3.970e-04 3.743e-04 5.246e-04 WNHE SFW WBSW CR 2.256e-04 3.121e-04 1.070e-03 3.795e-04 LK RWB AUR STTH 3.270e-04 5.362e-04 2.258e-04 3.030e-04 LR WPHE YTH ER 3.448e-04 1.088e-03 4.462e-04 4.023e-04 PCU ESP SCR RBFT 5.698e-04 1.020e-04 4.593e-04 9.532e-05 BFCS WAG WWCH NHHE 1.590e-04 4.435e-04 6.529e-04 4.579e-04 VS CST BTR AMAG 4.017e-04 3.738e-04 1.879e-04 2.768e-04 SCC RWH WSW STP 1.293e-04 4.836e-04 2.857e-04 3.781e-04 YFHE WHIP GAL FHE 2.499e-04 4.947e-04 2.709e-04 1.819e-04 BRTH SPP SIL GCU 2.000e-04 3.802e-04 1.871e-04 2.461e-04 MUSK MGLK BHHE RFC 2.261e-04 2.003e-04 4.002e-04 1.642e-04 YTBC LYRE CHE OWH 2.324e-04 3.176e-04 2.172e-04 1.111e-04 TRM MB STHR LHE 1.222e-04 3.187e-04 1.197e-04 1.715e-04 FTC PINK OBO YR 3.327e-04 1.348e-04 3.043e-04 1.495e-04 LFB SPW RBTR DWS 1.942e-04 2.064e-04 5.152e-04 3.397e-04 BELL LWB CBW GGC 1.934e-04 2.976e-04 4.610e-04 1.362e-04 PIL SKF RSL PDOV 2.051e-04 6.308e-04 1.241e-04 6.597e-05 CRP JW BCHE RCR 9.389e-05 1.429e-04 1.076e-04 1.860e-04 GBB RRP LLOR YTHE 4.313e-04 1.666e-04 5.725e-05 1.064e-04 RF SHBC AZKF SFC 3.168e-04 3.014e-04 1.955e-04 1.755e-04 YRTH ROSE BCOO LFC 1.275e-04 1.418e-04 1.150e-04 1.282e-04 WG PCOO WTG NMIN 1.180e-04 1.139e-04 2.769e-04 4.055e-04 NFB DB RBEE HBC 1.428e-04 2.607e-04 1.780e-04 2.634e-04 DF PCL FLAME WWT 1.462e-04 1.139e-04 1.570e-04 1.475e-04 WBWS LCOR KING 5.725e-05 6.195e-05 1.443e-04 - We now need to generate a distance matrix reflecting the multidimensional distance between each of the site pairs.
The Bray-Curtis dissimilarity index is a very popular distance metric as it:
- Reaches a maximum of 1 when two sites have nothing in common
- Reaches a minimum of 0 when two sites are identical
- Ignores shared absences, so dissimilarity is based only on the values of characteristics that the objects do share - two sites are not considered similar just because they both do not contain a certain species.
Show code> macnally.dist <- vegdist(macnally.std,"bray")
- Perform a non-metric multidimensional scaling with:
- two dimensions ($k=2$)
- use principle coordinates analysis to create the initial configuration
- use a maximum of 20 random starts
- use (modified) Kruskal's stress to determine the match between ordination distances and ecological distances
- use procrustes rotation to determine whether the configurations have converged
- center the axes scores and rotate the points such that the greatest plane of variation is orientated with the primary axis
Show code> macnally.mds <- metaMDS(macnally.dist, k=2)
Run 0 stress 0.1239 Run 1 stress 0.1239 ... procrustes: rmse 0.0005699 max resid 0.002667 *** Solution reached
- What is the final stress value?
Show code
> macnally.mds$stress
[1] 0.1239
- What does this stress value suggest about the success of the MDS, are two dimensions adequate?
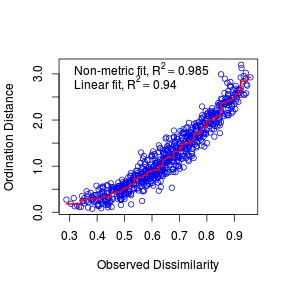
- The Sheppard diagram (stress plot) represents the relationship between the ecological distances (y-axis) and the new MDS ordination distances (x-axis).
How would you describe the shape of this curve, and base on this is metric or non-metric MDS more appropriate?
Show code> stressplot(macnally.mds)
-
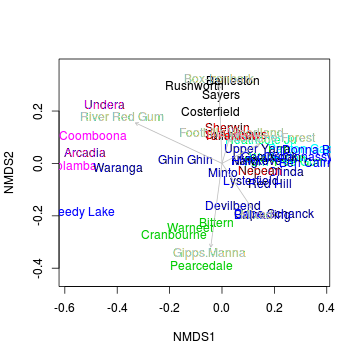
Generate an ordination plot to represent the similarity of the bird communities of the sites graphically.
Show code
> ordiplot(macnally.mds, display="sites", type="n") > text(macnally.mds,lab=rownames(macnally), col=as.numeric(macnally$HABITAT)) > habitat <- model.matrix(~-1+macnally$HABITAT) > colnames(habitat) <-gsub("macnally\\$HABITAT","",colnames(habitat)) > envfit <- envfit(macnally.mds, env=habitat) > envfit
***VECTORS NMDS1 NMDS2 r2 Box-Ironbark -0.038 0.999 0.35 Foothills Woodland 0.283 0.959 0.04 Gipps.Manna -0.133 -0.991 0.41 Mixed 0.554 -0.832 0.17 Montane Forest 0.888 0.460 0.10 River Red Gum -0.906 0.424 0.53 Pr(>r) Box-Ironbark 0.003 ** Foothills Woodland 0.529 Gipps.Manna 0.001 *** Mixed 0.051 . Montane Forest 0.143 River Red Gum 0.001 *** --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 P values based on 999 permutations.> plot(envfit, col="gray")
The nature of mathematics is that large numbers yield larger and more disparate values than smaller values. Hence in order to describe the communities in a way that all species and sites have an equal opportunity to influence the patterns, we should first standardize the species abundances.
There are a number of valid standardizations that we could attempt (and indeed you are encouraged to try a couple of alternatives to the one we use here). One popular and suitable standardization for ecological species abundance data is the Wisconsin double standardization. Not only does this standardization help suppress the dominance of the overly abundant species, it also magnifies the main patterns thereby making them easier to detect.
- Calculate the means and maximums of each species
