Workshop 9.2b - Nested designs (hierarchical models)
28 Jan 2015
Nested ANOVA (Mixed effects) references
- McCarthy (2007) - Chpt ?
- Kery (2010) - Chpt ?
- Gelman & Hill (2007) - Chpt ?
- Logan (2010) - Chpt 12
- Quinn & Keough (2002) - Chpt 9
Nested ANOVA - one between factor
In an unusually detailed preparation for an Environmental Effects Statement for a proposed discharge of dairy wastes into the Curdies River, in western Victoria, a team of stream ecologists wanted to describe the basic patterns of variation in a stream invertebrate thought to be sensitive to nutrient enrichment. As an indicator species, they focused on a small flatworm, Dugesia, and started by sampling populations of this worm at a range of scales. They sampled in two seasons, representing different flow regimes of the river - Winter and Summer. Within each season, they sampled three randomly chosen (well, haphazardly, because sites are nearly always chosen to be close to road access) sites. A total of six sites in all were visited, 3 in each season. At each site, they sampled six stones, and counted the number of flatworms on each stone.
Download Curdies data set| Format of curdies.csv data files | |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
 |
||||||||||||||||||||||||||||||||||||||||||||
The hierarchical nature of this design can be appreciated when we examine an illustration of the spatial and temporal scale of the measurements.
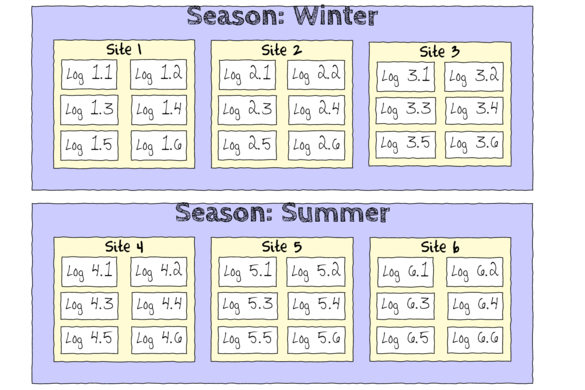
- Within each of the two Seasons, there were three separate Sites (Sites not repeatidly measured across Seasons).
- Within each of the Sites, six logs were selected (haphazardly) from which the number of flatworms were counted.
We construct the model in much the same way. That is:
- The observed number of flatworm per log (for a given Site) is equal to (modelled against) the mean number for that Site
plus a random amount drawn from a normal distribution with a mean of zero and a certain degree of variability (precision).
Number of flatwormlog i = βSite j,i + εi where ε ∼ N(0,σ2)
- The mean number of flatworm per Site (within a given Season) is equal to (modelled against) the mean number for that Season
plus a random amount drawn from a normal distribution with a means of zero and a certain degree of variability (precision).
μSite j = γSeason j + εj where ε ∼ N(0,σ2)
The SITE variable is conveniently already coded as a numeric series (starting at 1).
Open the curdies data file.curdies <- read.table('../downloads/data/curdies.csv', header=T, sep=',', strip.white=T) head(curdies)
SEASON SITE DUGESIA S4DUGES 1 WINTER 1 0.6477 0.8971 2 WINTER 1 6.0962 1.5713 3 WINTER 1 1.3106 1.0700 4 WINTER 1 1.7253 1.1461 5 WINTER 1 1.4594 1.0991 6 WINTER 1 1.0576 1.0141
- Lets start by performing some exploratory data analysis to help us identify any issues with the data as
well as helping to try to identify a possible model to fit.
- Boxplots for the main effect (Season)
Show codeIs there any evidence of non-normality and/or heterogeneity (Y or N)
library(dplyr) curdies.ag <- curdies %>% group_by(SEASON,SITE) %>% summarize(DUGESIA=mean(DUGESIA)) curdies.ag
Source: local data frame [6 x 3] Groups: SEASON SEASON SITE DUGESIA 1 SUMMER 4 0.4191 2 SUMMER 5 0.2291 3 SUMMER 6 0.1942 4 WINTER 1 2.0494 5 WINTER 2 4.1819 6 WINTER 3 0.6782
#OR equivalently with the plyr package library(plyr) ddply(curdies, ~SEASON+SITE, numcolwise(mean))
SEASON SITE DUGESIA S4DUGES 1 SUMMER 4 0.4191 0.3508 2 SUMMER 5 0.2291 0.1805 3 SUMMER 6 0.1942 0.3811 4 WINTER 1 2.0494 1.1329 5 WINTER 2 4.1819 1.2719 6 WINTER 3 0.6782 0.8679
detach(package:plyr) #since dplyr and plyr do not play nicely together library(ggplot2) ggplot(curdies.ag, aes(y=DUGESIA, x=SEASON)) + geom_boxplot()
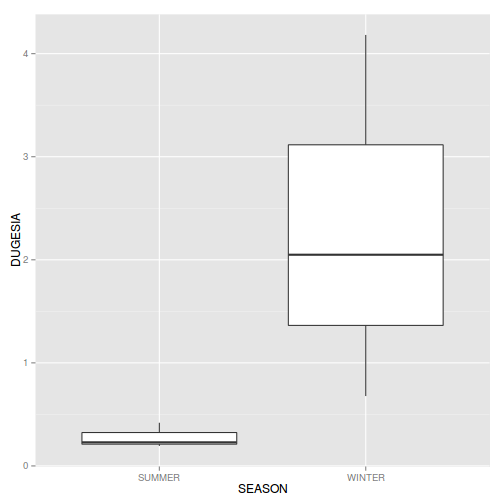
If so, assess whether a transformation (such as a forth-root transformation) will address the violations and then make the appropriate corrections - Consider a transformation (such as forth-root transform) and redo boxplots for the main effect (Season)
Show codeDoes a forth-root transformation improve the data with respect to normality and homogeneity of variance (Y or N)?
curdies.ag <- curdies %>% group_by(SEASON,SITE) %>% summarize(DUGESIA=mean(DUGESIA^0.25)) curdies.ag
Source: local data frame [6 x 3] Groups: SEASON SEASON SITE DUGESIA 1 SUMMER 4 0.3508 2 SUMMER 5 0.1805 3 SUMMER 6 0.3811 4 WINTER 1 1.1329 5 WINTER 2 1.2719 6 WINTER 3 0.8679
ggplot(curdies.ag, aes(y=DUGESIA, x=SEASON)) + geom_boxplot()
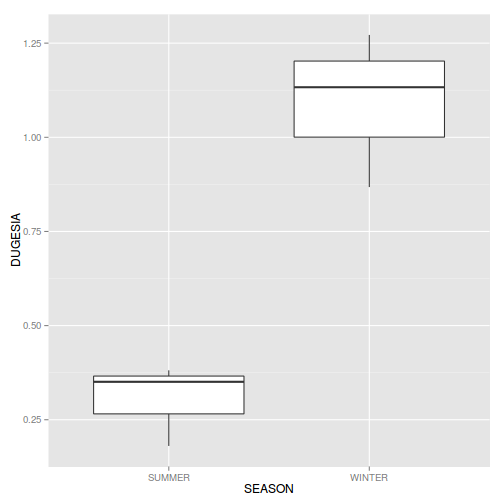
- Boxplots for the main effect (Season)
- Fit the hierarchical model.
Use with effects (or matrix) parameterization for the 'fixed effects' and means (or matrix) parameterization for the 'random effects'.
Note the following points:
- Non-informative priors for residual precision (standard deviations) should be defined as vague uniform distributions rather than gamma distributions. This prevents the chains from walking to close to parameter estimates of 0. Alternatively, Gelman recommends the use of weakly informative half-cauchy distributions for variances in hierarchical designs.
- Finite-population standard deviation for Site (the within group factor) should reflect standard deviation between Sites within season (not standard deviation between all Sites). Unfortunately, the BUGS language does not offer a mechanism to determine which indexes match certain criteria (for example which Sites correspond to which Season), nor does it allow variables to be redefined. One way to allow us to generate multiple standard deviations is to create an offset vector that can be used to indicate the range of Site numbers that are within which Season.
- As this is a multi-level design in which the Season 'replicates' are determined by the Site means, it is important that firstly, the number of levels of the within Group factor (Season) reflects this, and secondly that the factor levels of the within group factor (Season) correspond to the factor levels of the group factor (Site). In this case, the Sites are labeled 1-6, but the first three Sites are in Winter. R will naturally label Winter as the second level (as alphabetically W comes after S for summer). Thus without intervention or careful awareness, it will appear that the Winter means are higher than the Summer means, when it is actually the other way around. One way to check this, is create the data list and check that all the factor levels are listed in increasing numerical order.
View preparation codesites<-ddply(curdies, ~SITE, catcolwise(unique)) offset <- c(match(levels(sites$SEASON),sites$SEASON), length(sites$SEASON)+1) offset
[1] 4 1 7
curdies.list <- with(curdies, list(dugesia=DUGESIA, season=as.numeric(SEASON), site=as.numeric(SITE), n=nrow(curdies), nSeason=length(levels(as.factor(SEASON))), nSite=length(levels(as.factor(SITE))), SiteOffset=offset ) ) curdies.list
$dugesia [1] 0.6477 6.0962 1.3106 1.7253 1.4594 1.0576 1.0163 16.1968 1.1681 1.0243 2.0113 3.6746 [13] 0.6891 1.2191 1.1131 0.6569 0.1361 0.2547 0.0000 0.0000 0.9411 0.0000 0.0000 1.5735 [25] 1.3745 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1329 0.0000 0.6580 0.3745 $season [1] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 $site [1] 1 1 1 1 1 1 2 2 2 2 2 2 3 3 3 3 3 3 4 4 4 4 4 4 5 5 5 5 5 5 6 6 6 6 6 6 $n [1] 36 $nSeason [1] 2 $nSite [1] 6 $SiteOffset [1] 4 1 7
## notice that The Season variable has as many entries as there were plots ## rather than having as many as there were Sites. ## Also notice that Winter is the second Season according to the factor levels ## Finally notice that the offset values are not in order #season <- with(curdies,SEASON[match(unique(SITE),SITE)]) #season <- factor(season, levels=unique(season)) curdies$SEASON <- factor(curdies$SEASON, levels=c("WINTER","SUMMER")) sites<-ddply(curdies, ~SITE, catcolwise(unique)) offset <- c(match(levels(sites$SEASON),sites$SEASON), length(sites$SEASON)+1) offset
[1] 1 4 7
curdies.list <- with(curdies, list(dugesia=DUGESIA, season=as.numeric(SEASON), site=as.numeric(SITE), n=nrow(curdies), nSeason=length(levels(SEASON)), nSite=length(levels(as.factor(SITE))), SiteOffset=offset ) ) curdies.list
$dugesia [1] 0.6477 6.0962 1.3106 1.7253 1.4594 1.0576 1.0163 16.1968 1.1681 1.0243 2.0113 3.6746 [13] 0.6891 1.2191 1.1131 0.6569 0.1361 0.2547 0.0000 0.0000 0.9411 0.0000 0.0000 1.5735 [25] 1.3745 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1329 0.0000 0.6580 0.3745 $season [1] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 $site [1] 1 1 1 1 1 1 2 2 2 2 2 2 3 3 3 3 3 3 4 4 4 4 4 4 5 5 5 5 5 5 6 6 6 6 6 6 $n [1] 36 $nSeason [1] 2 $nSite [1] 6 $SiteOffset [1] 1 4 7
## That is betterShow full effects parameterization JAGS codemodelString=" model { #Likelihood for (i in 1:n) { y[i]~dnorm(mu[i],tau) mu[i] <- alpha0 + alpha[SEASON[i]] + beta[SITE[i]] y.err[i] <- y[i] - mu[i] } #Priors alpha0 ~ dnorm(0, 1.0E-6) alpha[1] <- 0 for (i in 2:nSEASON) { alpha[i] ~ dnorm(0, 1.0E-6) #prior } for (i in 1:nSITE) { beta[i] ~ dnorm(0, tau.B) #prior } #Uniform uninformative prior on variance tau <- 1 / (sigma * sigma) tau.B <- 1 / (sigma.B * sigma.B) sigma ~ dunif(0, 100) sigma.B ~ dunif(0, 100) for (i in 1:nSEASON) { SD.Site[i] <- sd(beta[SiteOffset[i]:SiteOffset[i+1]-1]) } sd.Season <- sd(alpha) sd.Site <- mean(SD.Site) #sd(beta.site) sd.Resid <- sd(y.err) } " writeLines(modelString,con="../downloads/BUGSscripts/ws9.2bS1.2a.txt") # Generate a data list curdies$SEASON <- factor(curdies$SEASON, levels=c("WINTER","SUMMER")) sites<-ddply(curdies, ~SITE, catcolwise(unique)) offset <- c(match(levels(sites$SEASON),sites$SEASON), length(sites$SEASON)+1) offset
[1] 1 4 7
curdies$SITE <- factor(curdies$SITE) curdies.list <- with(curdies, list(y=S4DUGES, SITE=as.numeric(SITE), SEASON=as.numeric(SEASON), n=nrow(curdies), nSITE=length(levels(SITE)), nSEASON = length(levels(SEASON)), SiteOffset=offset ) ) curdies.list
$y [1] 0.8971 1.5713 1.0700 1.1461 1.0991 1.0141 1.0040 2.0061 1.0396 1.0060 1.1909 1.3845 0.9111 [14] 1.0508 1.0272 0.9003 0.6074 0.7104 0.0000 0.0000 0.9849 0.0000 0.0000 1.1200 1.0828 0.0000 [27] 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.6038 0.0000 0.9007 0.7823 $SITE [1] 1 1 1 1 1 1 2 2 2 2 2 2 3 3 3 3 3 3 4 4 4 4 4 4 5 5 5 5 5 5 6 6 6 6 6 6 $SEASON [1] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 $n [1] 36 $nSITE [1] 6 $nSEASON [1] 2 $SiteOffset [1] 1 4 7
# define parameters params <- c("alpha0","alpha","sigma","sigma.B","sd.Season","sd.Site","sd.Resid","SD.Site") burnInSteps = 3000 nChains = 3 numSavedSteps = 3000 thinSteps = 10 nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) library(R2jags) curdies.r2jags.a <- jags(data=curdies.list, inits=NULL, parameters.to.save=params, model.file="../downloads/BUGSscripts/ws9.2bS1.2a.txt", n.chains=nChains, n.iter=nIter, n.burnin=burnInSteps, n.thin=thinSteps )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 176 Initializing model
print(curdies.r2jags.a)
Inference for Bugs model at "../downloads/BUGSscripts/ws9.2bS1.2a.txt", fit using jags, 3 chains, each with 13000 iterations (first 3000 discarded), n.thin = 10 n.sims = 3000 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff SD.Site[1] 0.119 0.092 0.004 0.043 0.101 0.177 0.340 1.008 370 SD.Site[2] 0.095 0.074 0.004 0.036 0.078 0.137 0.275 1.012 260 alpha[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 alpha[2] -0.785 0.242 -1.251 -0.915 -0.793 -0.661 -0.284 1.002 1600 alpha0 1.091 0.173 0.722 1.007 1.093 1.186 1.414 1.004 3000 sd.Resid 0.388 0.015 0.366 0.378 0.386 0.394 0.424 1.001 3000 sd.Season 0.557 0.165 0.211 0.468 0.561 0.647 0.885 1.006 2700 sd.Site 0.107 0.071 0.005 0.048 0.098 0.152 0.273 1.013 260 sigma 0.402 0.053 0.317 0.364 0.397 0.434 0.517 1.001 3000 sigma.B 0.176 0.159 0.006 0.067 0.134 0.235 0.606 1.009 350 deviance 34.995 3.529 29.652 32.541 34.456 36.830 43.240 1.001 3000 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 6.2 and DIC = 41.2 DIC is an estimate of expected predictive error (lower deviance is better).curdies.mcmc.a <- as.mcmc(curdies.r2jags.a)
Show matrix parameterization JAGS codemodelString=" model { #Likelihood for (i in 1:n) { y[i]~dnorm(mu[i],tau) mu[i] <- inprod(alpha[],X[i,]) + beta[SITE[i]] y.err[i] <- y[i] - mu[i] } #Priors alpha ~ dmnorm(a0, A0) for (i in 1:nSITE) { beta[i] ~ dnorm(0, tau.B) #prior } #Uniform uninformative prior on variance tau <- 1 / (sigma * sigma) tau.B <- 1 / (sigma.B * sigma.B) sigma ~ dunif(0, 100) sigma.B ~ dunif(0, 100) # Finite-population standard deviations for (i in 1:nSEASON) { SD.Site[i] <- sd(beta[SiteOffset[i]:SiteOffset[i+1]-1]) } seasons[1] <- alpha[1] for (i in 2:nSEASON) { seasons[i] <- alpha[1]+alpha[i] } sd.Season <- sd(seasons) sd.Site <- mean(SD.Site) sd.Resid <- sd(y.err) } " writeLines(modelString,con="../downloads/BUGSscripts/ws9.2bS1.2b.txt") # Generate a data list curdies$SEASON <- factor(curdies$SEASON, levels=c("WINTER","SUMMER")) sites<-ddply(curdies, ~SITE, catcolwise(unique)) offset <- c(match(levels(sites$SEASON),sites$SEASON), length(sites$SEASON)+1) offset
[1] 1 4 7
curdies$SITE <- factor(curdies$SITE) Xmat <- model.matrix(~SEASON, data=curdies) curdies.list <- with(curdies, list(y=S4DUGES, SITE=as.numeric(SITE), X=Xmat, n=nrow(curdies), nSITE=length(levels(SITE)), nSEASON=length(levels(SEASON)), a0=rep(0, ncol(Xmat)), A0=diag(0, ncol(Xmat)), SiteOffset=offset ) ) curdies.list
$y [1] 0.8971 1.5713 1.0700 1.1461 1.0991 1.0141 1.0040 2.0061 1.0396 1.0060 1.1909 1.3845 0.9111 [14] 1.0508 1.0272 0.9003 0.6074 0.7104 0.0000 0.0000 0.9849 0.0000 0.0000 1.1200 1.0828 0.0000 [27] 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.6038 0.0000 0.9007 0.7823 $SITE [1] 1 1 1 1 1 1 2 2 2 2 2 2 3 3 3 3 3 3 4 4 4 4 4 4 5 5 5 5 5 5 6 6 6 6 6 6 $X (Intercept) SEASONSUMMER 1 1 0 2 1 0 3 1 0 4 1 0 5 1 0 6 1 0 7 1 0 8 1 0 9 1 0 10 1 0 11 1 0 12 1 0 13 1 0 14 1 0 15 1 0 16 1 0 17 1 0 18 1 0 19 1 1 20 1 1 21 1 1 22 1 1 23 1 1 24 1 1 25 1 1 26 1 1 27 1 1 28 1 1 29 1 1 30 1 1 31 1 1 32 1 1 33 1 1 34 1 1 35 1 1 36 1 1 attr(,"assign") [1] 0 1 attr(,"contrasts") attr(,"contrasts")$SEASON [1] "contr.treatment" $n [1] 36 $nSITE [1] 6 $nSEASON [1] 2 $a0 [1] 0 0 $A0 [,1] [,2] [1,] 0 0 [2,] 0 0 $SiteOffset [1] 1 4 7# define parameters params <- c("alpha","sigma","sigma.B","sd.Season","sd.Site","sd.Resid","SD.Site") burnInSteps = 3000 nChains = 3 numSavedSteps = 3000 thinSteps = 10 nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) library(R2jags) curdies.r2jags.b <- jags(data=curdies.list, inits=NULL, parameters.to.save=params, model.file="../downloads/BUGSscripts/ws9.2bS1.2b.txt", n.chains=nChains, n.iter=nIter, n.burnin=burnInSteps, n.thin=thinSteps )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 258 Initializing model
print(curdies.r2jags.b)
Inference for Bugs model at "../downloads/BUGSscripts/ws9.2bS1.2b.txt", fit using jags, 3 chains, each with 13000 iterations (first 3000 discarded), n.thin = 10 n.sims = 3000 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff SD.Site[1] 0.118 0.090 0.005 0.044 0.099 0.174 0.328 1.009 320 SD.Site[2] 0.094 0.072 0.004 0.037 0.078 0.134 0.270 1.012 370 alpha[1] 1.090 0.171 0.735 1.001 1.090 1.178 1.431 1.002 3000 alpha[2] -0.785 0.234 -1.225 -0.908 -0.786 -0.663 -0.323 1.001 3000 sd.Resid 0.388 0.015 0.366 0.378 0.386 0.394 0.424 1.001 3000 sd.Season 0.556 0.160 0.232 0.469 0.556 0.642 0.866 1.016 1300 sd.Site 0.106 0.070 0.006 0.051 0.096 0.151 0.263 1.014 290 sigma 0.403 0.051 0.317 0.367 0.398 0.434 0.518 1.001 3000 sigma.B 0.171 0.158 0.007 0.064 0.132 0.226 0.606 1.014 210 deviance 34.970 3.342 29.803 32.685 34.584 36.780 42.822 1.001 3000 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 5.6 and DIC = 40.6 DIC is an estimate of expected predictive error (lower deviance is better).curdies.mcmc.b <- as.mcmc(curdies.r2jags.b)
Hierarchical parameterization JAGS codemodelString=" model { #Likelihood #Likelihood (esimating site means (gamma.site) for (i in 1:n) { y[i]~dnorm(quad.means[i],tau) quad.means[i] <- gamma.site[SITE[i]] y.err[i] <- y[i] - quad.means[i] } for (i in 1:nSITE) { gamma.site[i] ~ dnorm(site.means[i], tau.site) site.means[i] <- inprod(beta[],X[i,]) } #Priors for (i in 1:nX) { beta[i] ~ dnorm(0, 1.0E-6) #prior } sigma ~ dunif(0, 100) tau <- 1 / (sigma * sigma) sigma.site ~ dunif(0, 100) tau.site <- 1 / (sigma.site * sigma.site) # Finite-population standard deviations for (i in 1:nSEASON) { SD.Site[i] <- sd(gamma.site[SiteOffset[i]:SiteOffset[i+1]-1]) } seasons[1] <- beta[1] for (i in 2:nSEASON) { seasons[i] <- beta[1]+beta[i] } sd.Season <- sd(seasons) sd.Site <- mean(SD.Site) sd.Resid <- sd(y.err) } " writeLines(modelString,con="../downloads/BUGSscripts/ws9.2bS1.2c.txt") # Generate a data list curdies$SEASON <- factor(curdies$SEASON, levels=c("WINTER","SUMMER")) sites<-ddply(curdies, ~SITE, catcolwise(unique)) offset <- c(match(levels(sites$SEASON),sites$SEASON), length(sites$SEASON)+1) offset
[1] 1 4 7
curdies$SITE <- factor(curdies$SITE) Xmat <- model.matrix(~SEASON, data=ddply(curdies, ~SITE, catcolwise(unique))) curdies.list <- with(curdies, list(y=S4DUGES, SITE=as.numeric(SITE), X=Xmat, nX=ncol(Xmat), n=nrow(curdies), nSITE=length(levels(SITE)), nSEASON=length(levels(SEASON)), SiteOffset=offset ) ) curdies.list
$y [1] 0.8971 1.5713 1.0700 1.1461 1.0991 1.0141 1.0040 2.0061 1.0396 1.0060 1.1909 1.3845 0.9111 [14] 1.0508 1.0272 0.9003 0.6074 0.7104 0.0000 0.0000 0.9849 0.0000 0.0000 1.1200 1.0828 0.0000 [27] 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.6038 0.0000 0.9007 0.7823 $SITE [1] 1 1 1 1 1 1 2 2 2 2 2 2 3 3 3 3 3 3 4 4 4 4 4 4 5 5 5 5 5 5 6 6 6 6 6 6 $X (Intercept) SEASONSUMMER 1 1 0 2 1 0 3 1 0 4 1 1 5 1 1 6 1 1 attr(,"assign") [1] 0 1 attr(,"contrasts") attr(,"contrasts")$SEASON [1] "contr.treatment" $nX [1] 2 $n [1] 36 $nSITE [1] 6 $nSEASON [1] 2 $SiteOffset [1] 1 4 7
# define parameters params <- c("beta","sigma","sigma.site","sd.Season","sd.Site","sd.Resid") burnInSteps = 3000 nChains = 3 numSavedSteps = 3000 thinSteps = 10 nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) library(R2jags) curdies.r2jags.c <- jags(data=curdies.list, inits=NULL, parameters.to.save=params, model.file="../downloads/BUGSscripts/ws9.2bS1.2c.txt", n.chains=nChains, n.iter=nIter, n.burnin=burnInSteps, n.thin=thinSteps )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 156 Initializing model
print(curdies.r2jags.c)
Inference for Bugs model at "../downloads/BUGSscripts/ws9.2bS1.2c.txt", fit using jags, 3 chains, each with 13000 iterations (first 3000 discarded), n.thin = 10 n.sims = 3000 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff beta[1] 1.092 0.161 0.784 1.003 1.090 1.177 1.418 1.001 3000 beta[2] -0.791 0.227 -1.247 -0.913 -0.789 -0.666 -0.344 1.001 3000 sd.Resid 0.387 0.014 0.366 0.378 0.386 0.394 0.422 1.001 3000 sd.Season 0.561 0.155 0.246 0.471 0.558 0.646 0.882 1.002 3000 sd.Site 0.103 0.070 0.005 0.046 0.094 0.149 0.259 1.005 530 sigma 0.403 0.054 0.314 0.367 0.396 0.436 0.525 1.002 1200 sigma.site 0.162 0.146 0.006 0.062 0.127 0.217 0.540 1.003 720 deviance 35.040 3.427 29.731 32.680 34.510 36.887 43.062 1.001 3000 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 5.9 and DIC = 40.9 DIC is an estimate of expected predictive error (lower deviance is better).curdies.mcmc.c <- as.mcmc(curdies.r2jags.c)
Matrix parameterization STAN codeUnfortunately, STAN does not yet support vector indexing or the ability to multiply a matrix by a vector. A work around involves multiplication of a dummy matrix with the gamma vector..rstanString= # Generate a data list curdies$SEASON <- factor(curdies$SEASON, levels=c("WINTER","SUMMER")) sites<-ddply(curdies, ~SITE, catcolwise(unique)) offset <- c(match(levels(sites$SEASON),sites$SEASON), length(sites$SEASON)+1) offset
[1] 1 4 7
curdies$SITE <- factor(curdies$SITE) Xmat <- model.matrix(~SEASON, data=curdies) Zmat <- model.matrix(~-1+SITE, data=curdies) curdies.list <- with(curdies, list(y=S4DUGES, Z=Zmat, X=Xmat, nX=ncol(Xmat), nZ=ncol(Zmat), n=nrow(curdies), a0=rep(0,ncol(Xmat)), A0=diag(100000,ncol(Xmat)), SiteOffset=offset, m=matrix(rep(1,6),2) ) ) curdies.list
$y [1] 0.8971 1.5713 1.0700 1.1461 1.0991 1.0141 1.0040 2.0061 1.0396 1.0060 1.1909 1.3845 0.9111 [14] 1.0508 1.0272 0.9003 0.6074 0.7104 0.0000 0.0000 0.9849 0.0000 0.0000 1.1200 1.0828 0.0000 [27] 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.6038 0.0000 0.9007 0.7823 $Z SITE1 SITE2 SITE3 SITE4 SITE5 SITE6 1 1 0 0 0 0 0 2 1 0 0 0 0 0 3 1 0 0 0 0 0 4 1 0 0 0 0 0 5 1 0 0 0 0 0 6 1 0 0 0 0 0 7 0 1 0 0 0 0 8 0 1 0 0 0 0 9 0 1 0 0 0 0 10 0 1 0 0 0 0 11 0 1 0 0 0 0 12 0 1 0 0 0 0 13 0 0 1 0 0 0 14 0 0 1 0 0 0 15 0 0 1 0 0 0 16 0 0 1 0 0 0 17 0 0 1 0 0 0 18 0 0 1 0 0 0 19 0 0 0 1 0 0 20 0 0 0 1 0 0 21 0 0 0 1 0 0 22 0 0 0 1 0 0 23 0 0 0 1 0 0 24 0 0 0 1 0 0 25 0 0 0 0 1 0 26 0 0 0 0 1 0 27 0 0 0 0 1 0 28 0 0 0 0 1 0 29 0 0 0 0 1 0 30 0 0 0 0 1 0 31 0 0 0 0 0 1 32 0 0 0 0 0 1 33 0 0 0 0 0 1 34 0 0 0 0 0 1 35 0 0 0 0 0 1 36 0 0 0 0 0 1 attr(,"assign") [1] 1 1 1 1 1 1 attr(,"contrasts") attr(,"contrasts")$SITE [1] "contr.treatment" $X (Intercept) SEASONSUMMER 1 1 0 2 1 0 3 1 0 4 1 0 5 1 0 6 1 0 7 1 0 8 1 0 9 1 0 10 1 0 11 1 0 12 1 0 13 1 0 14 1 0 15 1 0 16 1 0 17 1 0 18 1 0 19 1 1 20 1 1 21 1 1 22 1 1 23 1 1 24 1 1 25 1 1 26 1 1 27 1 1 28 1 1 29 1 1 30 1 1 31 1 1 32 1 1 33 1 1 34 1 1 35 1 1 36 1 1 attr(,"assign") [1] 0 1 attr(,"contrasts") attr(,"contrasts")$SEASON [1] "contr.treatment" $nX [1] 2 $nZ [1] 6 $n [1] 36 $a0 [1] 0 0 $A0 [,1] [,2] [1,] 1e+05 0e+00 [2,] 0e+00 1e+05 $SiteOffset [1] 1 4 7 $m [,1] [,2] [,3] [1,] 1 1 1 [2,] 1 1 1# define parameters burnInSteps = 3000 nChains = 3 numSavedSteps = 3000 thinSteps = 10 nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) library(rstan) curdies.rstan.a <- stan(data=curdies.list, model_code=rstanString, pars=c('beta','gamma','sigma','sigma_Z','sd_Resid','sd_Season','sd_Site'), chains=nChains, iter=nIter, warmup=burnInSteps, thin=thinSteps, save_dso=TRUE )
TRANSLATING MODEL 'rstanString' FROM Stan CODE TO C++ CODE NOW. COMPILING THE C++ CODE FOR MODEL 'rstanString' NOW. SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 1). Iteration: 1 / 13000 [ 0%] (Warmup) Iteration: 1300 / 13000 [ 10%] (Warmup) Iteration: 2600 / 13000 [ 20%] (Warmup) Iteration: 3001 / 13000 [ 23%] (Sampling) Iteration: 4300 / 13000 [ 33%] (Sampling) Iteration: 5600 / 13000 [ 43%] (Sampling) Iteration: 6900 / 13000 [ 53%] (Sampling) Iteration: 8200 / 13000 [ 63%] (Sampling) Iteration: 9500 / 13000 [ 73%] (Sampling) Iteration: 10800 / 13000 [ 83%] (Sampling) Iteration: 12100 / 13000 [ 93%] (Sampling) Iteration: 13000 / 13000 [100%] (Sampling) # Elapsed Time: 0.33 seconds (Warm-up) # 1.21 seconds (Sampling) # 1.54 seconds (Total) SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 2). Iteration: 1 / 13000 [ 0%] (Warmup) Iteration: 1300 / 13000 [ 10%] (Warmup) Iteration: 2600 / 13000 [ 20%] (Warmup) Iteration: 3001 / 13000 [ 23%] (Sampling) Iteration: 4300 / 13000 [ 33%] (Sampling) Iteration: 5600 / 13000 [ 43%] (Sampling) Iteration: 6900 / 13000 [ 53%] (Sampling) Iteration: 8200 / 13000 [ 63%] (Sampling) Iteration: 9500 / 13000 [ 73%] (Sampling) Iteration: 10800 / 13000 [ 83%] (Sampling) Iteration: 12100 / 13000 [ 93%] (Sampling) Iteration: 13000 / 13000 [100%] (Sampling) # Elapsed Time: 0.36 seconds (Warm-up) # 1.13 seconds (Sampling) # 1.49 seconds (Total) SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 3). Iteration: 1 / 13000 [ 0%] (Warmup) Iteration: 1300 / 13000 [ 10%] (Warmup) Iteration: 2600 / 13000 [ 20%] (Warmup) Iteration: 3001 / 13000 [ 23%] (Sampling) Iteration: 4300 / 13000 [ 33%] (Sampling) Iteration: 5600 / 13000 [ 43%] (Sampling) Iteration: 6900 / 13000 [ 53%] (Sampling) Iteration: 8200 / 13000 [ 63%] (Sampling) Iteration: 9500 / 13000 [ 73%] (Sampling) Iteration: 10800 / 13000 [ 83%] (Sampling) Iteration: 12100 / 13000 [ 93%] (Sampling) Iteration: 13000 / 13000 [100%] (Sampling) # Elapsed Time: 0.33 seconds (Warm-up) # 1.74 seconds (Sampling) # 2.07 seconds (Total)
print(curdies.rstan.a)
Inference for Stan model: rstanString. 3 chains, each with iter=13000; warmup=3000; thin=10; post-warmup draws per chain=1000, total post-warmup draws=3000. mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat beta[1] 1.09 0.00 0.21 0.74 1.00 1.08 1.18 1.45 1922 1 beta[2] -0.78 0.01 0.30 -1.28 -0.91 -0.79 -0.66 -0.30 1905 1 gamma[1] 0.03 0.00 0.22 -0.34 -0.05 0.01 0.09 0.41 1939 1 gamma[2] 0.09 0.01 0.22 -0.22 -0.01 0.06 0.16 0.52 1737 1 gamma[3] -0.10 0.00 0.21 -0.52 -0.18 -0.07 0.00 0.18 1963 1 gamma[4] 0.02 0.00 0.20 -0.34 -0.05 0.01 0.10 0.43 2674 1 gamma[5] -0.06 0.00 0.19 -0.47 -0.12 -0.04 0.03 0.26 2363 1 gamma[6] 0.04 0.00 0.19 -0.33 -0.04 0.02 0.11 0.42 2879 1 sigma 0.40 0.00 0.05 0.32 0.36 0.40 0.43 0.52 2686 1 sigma_Z 0.20 0.01 0.23 0.03 0.08 0.15 0.25 0.69 1416 1 sd_Resid 0.39 0.00 0.02 0.37 0.38 0.38 0.39 0.43 2916 1 sd_Season 0.56 0.00 0.18 0.22 0.47 0.56 0.64 0.91 2494 1 sd_Site 0.12 0.00 0.07 0.02 0.06 0.11 0.16 0.27 1763 1 lp__ 22.11 0.12 4.35 13.11 19.41 22.13 24.95 30.61 1373 1 Samples were drawn using NUTS(diag_e) at Wed Jan 28 09:13:29 2015. For each parameter, n_eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence, Rhat=1).curdies.mcmc.d <- rstan:::as.mcmc.list.stanfit(curdies.rstan.a) curdies.mcmc.df.d <- as.data.frame(extract(curdies.rstan.a)) SD.site <- NULL sd.site <- curdies.mcmc.df.d[,3:8] for (i in 1:length(levels(curdies$SEASON))) { SD.site <- c(SD.site,apply(sd.site[,offset[i]:(offset[i+1]-1)],1,sd)) } data.frame(mean=mean(SD.site), median=median(SD.site), HPDinterval(as.mcmc(SD.site)), HPDinterval(as.mcmc(SD.site),p=0.68))
mean median lower upper lower.1 upper.1 var1 0.116 0.09714 0.002386 0.2794 0.0052 0.1446
Hierarchical parameterization STAN coderstanString=" data{ int n; int nZ; int nX; vector [n] y; matrix [nZ,nX] X; matrix [n,nZ] Z; } parameters{ vector [nX] beta; real<lower=0> sigma; vector [nZ] gamma; real<lower=0> sigma_Z; } transformed parameters { vector [n] mu_Z; vector [nZ] mu; mu_Z <- Z*gamma; mu <- X*beta; } model{ // Priors beta ~ normal(0, 10000); gamma ~ normal( 0 , 1000); sigma_Z ~ uniform( 0 , 100 ); sigma ~ uniform( 0 , 100 ); y ~ normal(mu_Z, sigma); gamma ~ normal( mu , sigma_Z ); } generated quantities { vector [n] y_err; real<lower=0> sd_Resid; y_err <- y - mu_Z; sd_Resid <- sd(y_err); } " # Generate a data list curdies$SITE <- factor(curdies$SITE) Xmat <- model.matrix(~SEASON, data=ddply(curdies, ~SITE, catcolwise(unique))) Zmat <- model.matrix(~-1+SITE, data=curdies) curdies.list <- with(curdies, list(y=S4DUGES, Z=Zmat, X=Xmat, nX=ncol(Xmat), nZ=ncol(Zmat), n=nrow(curdies) ) ) curdies.list
$y [1] 0.8971 1.5713 1.0700 1.1461 1.0991 1.0141 1.0040 2.0061 1.0396 1.0060 1.1909 1.3845 0.9111 [14] 1.0508 1.0272 0.9003 0.6074 0.7104 0.0000 0.0000 0.9849 0.0000 0.0000 1.1200 1.0828 0.0000 [27] 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.6038 0.0000 0.9007 0.7823 $Z SITE1 SITE2 SITE3 SITE4 SITE5 SITE6 1 1 0 0 0 0 0 2 1 0 0 0 0 0 3 1 0 0 0 0 0 4 1 0 0 0 0 0 5 1 0 0 0 0 0 6 1 0 0 0 0 0 7 0 1 0 0 0 0 8 0 1 0 0 0 0 9 0 1 0 0 0 0 10 0 1 0 0 0 0 11 0 1 0 0 0 0 12 0 1 0 0 0 0 13 0 0 1 0 0 0 14 0 0 1 0 0 0 15 0 0 1 0 0 0 16 0 0 1 0 0 0 17 0 0 1 0 0 0 18 0 0 1 0 0 0 19 0 0 0 1 0 0 20 0 0 0 1 0 0 21 0 0 0 1 0 0 22 0 0 0 1 0 0 23 0 0 0 1 0 0 24 0 0 0 1 0 0 25 0 0 0 0 1 0 26 0 0 0 0 1 0 27 0 0 0 0 1 0 28 0 0 0 0 1 0 29 0 0 0 0 1 0 30 0 0 0 0 1 0 31 0 0 0 0 0 1 32 0 0 0 0 0 1 33 0 0 0 0 0 1 34 0 0 0 0 0 1 35 0 0 0 0 0 1 36 0 0 0 0 0 1 attr(,"assign") [1] 1 1 1 1 1 1 attr(,"contrasts") attr(,"contrasts")$SITE [1] "contr.treatment" $X (Intercept) SEASONSUMMER 1 1 0 2 1 0 3 1 0 4 1 1 5 1 1 6 1 1 attr(,"assign") [1] 0 1 attr(,"contrasts") attr(,"contrasts")$SEASON [1] "contr.treatment" $nX [1] 2 $nZ [1] 6 $n [1] 36
# define parameters burnInSteps = 3000 nChains = 3 numSavedSteps = 3000 thinSteps = 10 nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) library(rstan) curdies.rstan.b <- stan(data=curdies.list, model_code=rstanString, pars=c('beta','gamma','sigma','sigma_Z','sd_Resid'), chains=nChains, iter=nIter, warmup=burnInSteps, thin=thinSteps, save_dso=TRUE )
TRANSLATING MODEL 'rstanString' FROM Stan CODE TO C++ CODE NOW. COMPILING THE C++ CODE FOR MODEL 'rstanString' NOW. SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 1). Iteration: 1 / 13000 [ 0%] (Warmup) Iteration: 1300 / 13000 [ 10%] (Warmup) Iteration: 2600 / 13000 [ 20%] (Warmup) Iteration: 3001 / 13000 [ 23%] (Sampling) Iteration: 4300 / 13000 [ 33%] (Sampling) Iteration: 5600 / 13000 [ 43%] (Sampling) Iteration: 6900 / 13000 [ 53%] (Sampling) Iteration: 8200 / 13000 [ 63%] (Sampling) Iteration: 9500 / 13000 [ 73%] (Sampling) Iteration: 10800 / 13000 [ 83%] (Sampling) Iteration: 12100 / 13000 [ 93%] (Sampling) Iteration: 13000 / 13000 [100%] (Sampling) # Elapsed Time: 0.31 seconds (Warm-up) # 0.88 seconds (Sampling) # 1.19 seconds (Total) SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 2). Iteration: 1 / 13000 [ 0%] (Warmup) Iteration: 1300 / 13000 [ 10%] (Warmup) Iteration: 2600 / 13000 [ 20%] (Warmup) Iteration: 3001 / 13000 [ 23%] (Sampling) Iteration: 4300 / 13000 [ 33%] (Sampling) Iteration: 5600 / 13000 [ 43%] (Sampling) Iteration: 6900 / 13000 [ 53%] (Sampling) Iteration: 8200 / 13000 [ 63%] (Sampling) Iteration: 9500 / 13000 [ 73%] (Sampling) Iteration: 10800 / 13000 [ 83%] (Sampling) Iteration: 12100 / 13000 [ 93%] (Sampling) Iteration: 13000 / 13000 [100%] (Sampling) # Elapsed Time: 0.35 seconds (Warm-up) # 1.62 seconds (Sampling) # 1.97 seconds (Total) SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 3). Iteration: 1 / 13000 [ 0%] (Warmup) Iteration: 1300 / 13000 [ 10%] (Warmup) Iteration: 2600 / 13000 [ 20%] (Warmup) Iteration: 3001 / 13000 [ 23%] (Sampling) Iteration: 4300 / 13000 [ 33%] (Sampling) Iteration: 5600 / 13000 [ 43%] (Sampling) Iteration: 6900 / 13000 [ 53%] (Sampling) Iteration: 8200 / 13000 [ 63%] (Sampling) Iteration: 9500 / 13000 [ 73%] (Sampling) Iteration: 10800 / 13000 [ 83%] (Sampling) Iteration: 12100 / 13000 [ 93%] (Sampling) Iteration: 13000 / 13000 [100%] (Sampling) # Elapsed Time: 0.33 seconds (Warm-up) # 0.89 seconds (Sampling) # 1.22 seconds (Total)
print(curdies.rstan.b)
Inference for Stan model: rstanString. 3 chains, each with iter=13000; warmup=3000; thin=10; post-warmup draws per chain=1000, total post-warmup draws=3000. mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat beta[1] 1.10 0.00 0.18 0.76 1.01 1.10 1.20 1.42 3000 1.01 beta[2] -0.79 0.00 0.26 -1.23 -0.91 -0.81 -0.67 -0.32 2856 1.00 gamma[1] 1.12 0.00 0.13 0.86 1.03 1.12 1.20 1.38 1001 1.01 gamma[2] 1.17 0.00 0.14 0.92 1.08 1.18 1.26 1.46 1475 1.00 gamma[3] 1.01 0.02 0.15 0.70 0.91 1.01 1.11 1.27 37 1.05 gamma[4] 0.33 0.00 0.13 0.07 0.24 0.33 0.40 0.59 1072 1.00 gamma[5] 0.25 0.01 0.14 -0.04 0.16 0.26 0.36 0.50 134 1.02 gamma[6] 0.34 0.00 0.13 0.08 0.25 0.35 0.42 0.61 1156 1.00 sigma 0.41 0.00 0.05 0.32 0.37 0.40 0.45 0.52 203 1.01 sigma_Z 0.18 0.01 0.20 0.02 0.07 0.13 0.23 0.64 206 1.01 sd_Resid 0.39 0.00 0.01 0.37 0.38 0.39 0.39 0.43 2219 1.00 lp__ 22.74 0.89 4.71 13.74 19.72 22.52 25.37 33.06 28 1.08 Samples were drawn using NUTS(diag_e) at Wed Jan 28 13:31:01 2015. For each parameter, n_eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence, Rhat=1).curdies.mcmc.e <- rstan:::as.mcmc.list.stanfit(curdies.rstan.b) curdies.mcmc.df.e <- as.data.frame(extract(curdies.rstan.b))
#Finite-population standard deviations ## Seasons seasons <- NULL seasons <- cbind(curdies.mcmc.df.e[,'beta.1'], curdies.mcmc.df.e[,'beta.1']+curdies.mcmc.df.e[,'beta.2']) sd.season <- apply(seasons,1,sd) library(coda) data.frame(mean=mean(sd.season), median=median(sd.season), HPDinterval(as.mcmc(sd.season)), HPDinterval(as.mcmc(sd.season),p=0.68))
mean median lower upper lower.1 upper.1 var1 0.5613 0.5727 0.2267 0.8611 0.4322 0.6935
## Sites SD.site <- NULL sd.site <- curdies.mcmc.df.e[,3:8] for (i in 1:length(levels(curdies$SEASON))) { SD.site <- c(SD.site,apply(sd.site[,offset[i]:(offset[i+1]-1)],1,sd)) } data.frame(mean=mean(SD.site), median=median(SD.site), HPDinterval(as.mcmc(SD.site)), HPDinterval(as.mcmc(SD.site),p=0.68))
mean median lower upper lower.1 upper.1 var1 0.1066 0.0864 0.001663 0.2648 0.007917 0.1357
- Before fully exploring the parameters, it is prudent to examine the convergence and mixing diagnostics.
Chose either any of the parameterizations (they should yield much the same) - although it is sometimes useful to explore the
different performances of effects vs matrix and JAGS vs STAN.
Full effects parameterization JAGS code
preds <- c("alpha0", "alpha[2]", "sigma", "sigma.B") plot(curdies.mcmc.a)
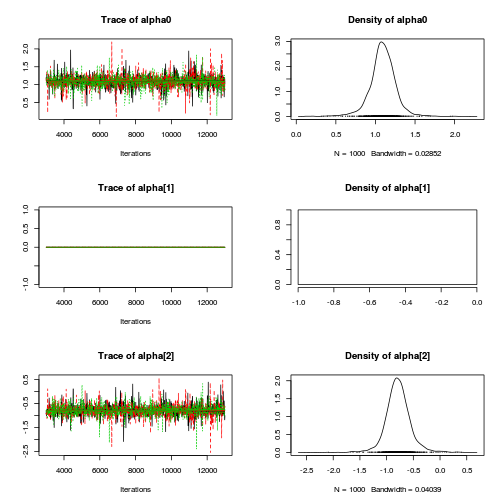
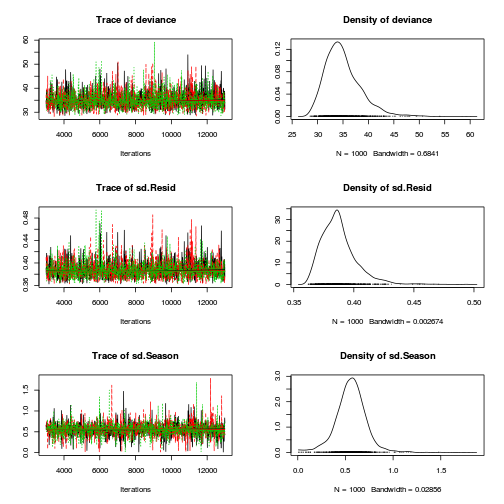
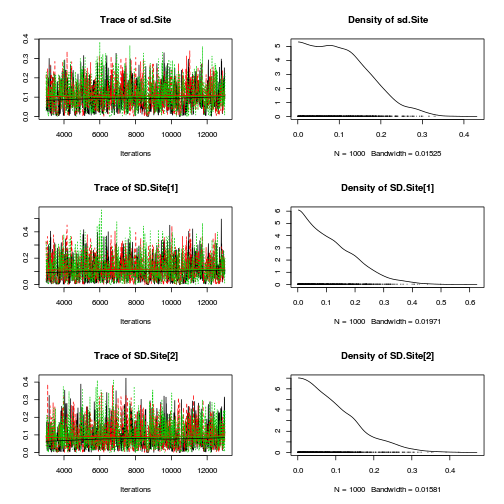
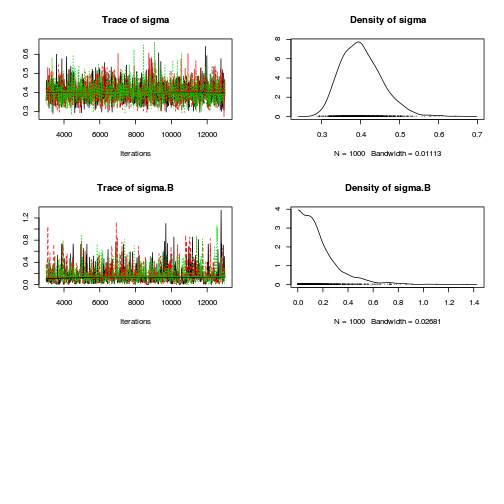
autocorr.diag(curdies.mcmc.a[, preds])
alpha0 alpha[2] sigma sigma.B Lag 0 1.000000 1.00000 1.00000 1.000000 Lag 10 0.002572 -0.04020 -0.01594 0.456169 Lag 50 0.003679 -0.01388 -0.01541 0.043737 Lag 100 0.002851 0.01658 0.01728 0.005163 Lag 500 0.033089 0.01574 -0.01057 0.007020
raftery.diag(curdies.mcmc.a)
[[1]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[2]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[3]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
Matrix parameterization JAGS codepreds <- c("alpha[1]", "alpha[2]", "sigma", "sigma.B") plot(curdies.mcmc.b)
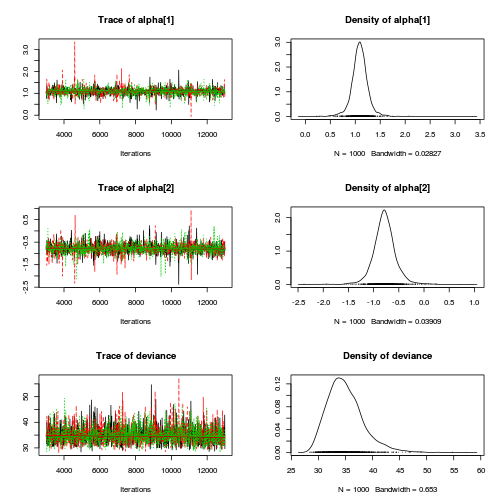
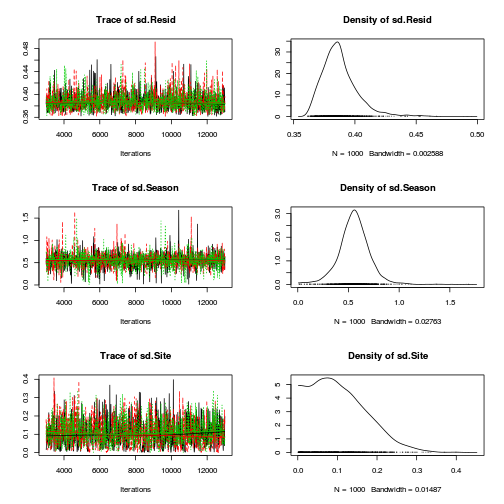
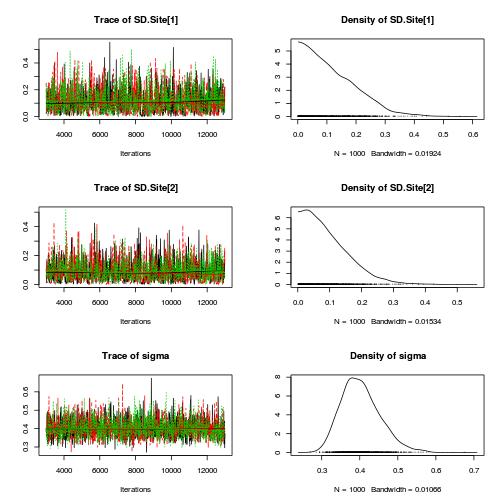
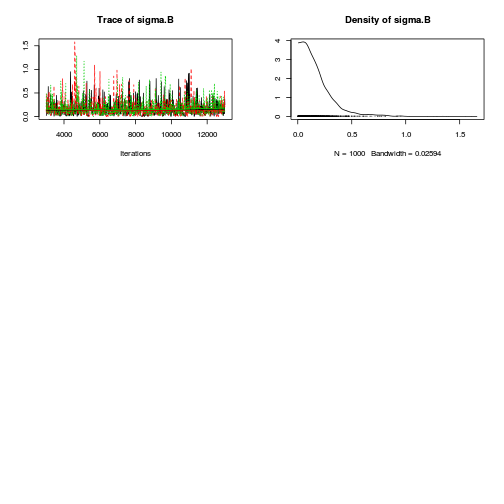
autocorr.diag(curdies.mcmc.b[, preds])
alpha[1] alpha[2] sigma sigma.B Lag 0 1.000000 1.00000 1.0000000 1.000000 Lag 10 0.042932 -0.02434 -0.0242340 0.432361 Lag 50 0.011679 -0.03564 0.0155186 0.043447 Lag 100 -0.048408 -0.03027 -0.0007029 -0.012678 Lag 500 0.001623 0.01541 0.0322009 0.006364
raftery.diag(curdies.mcmc.b)
[[1]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[2]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[3]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
Hierarchical parameterization JAGS codepreds <- c("beta[1]", "beta[2]", "sigma", "sigma.site") plot(curdies.mcmc.c)
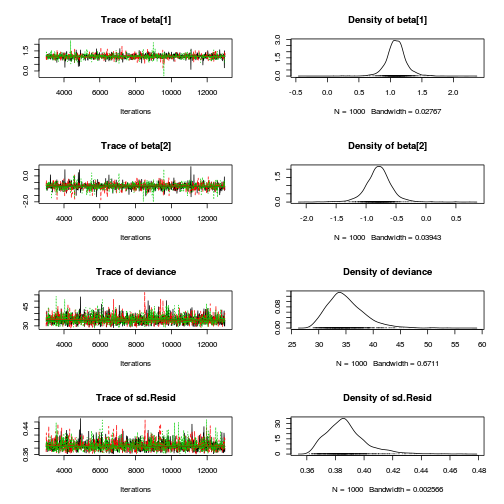
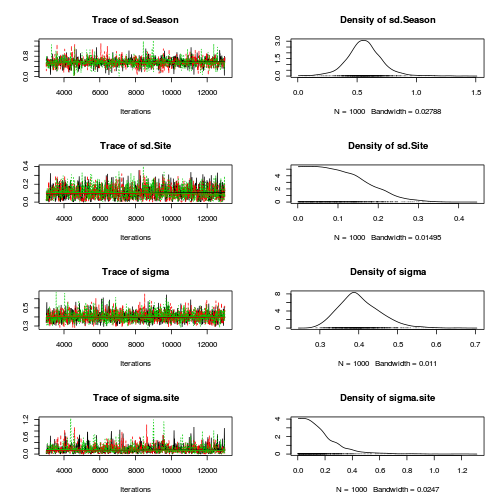
autocorr.diag(curdies.mcmc.c[, preds])
beta[1] beta[2] sigma sigma.site Lag 0 1.000000 1.000000 1.000000 1.000000 Lag 10 0.030968 0.073360 0.004006 0.400666 Lag 50 -0.004717 0.004116 0.006822 0.022614 Lag 100 -0.004866 0.010516 0.018844 -0.001234 Lag 500 -0.011233 -0.020237 -0.021071 0.010194
raftery.diag(curdies.mcmc.c)
[[1]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[2]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[3]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
Matrix parameterization STAN codepreds <- c("beta[1]", "beta[2]", "sigma", "sigma_Z") plot(curdies.mcmc.d)
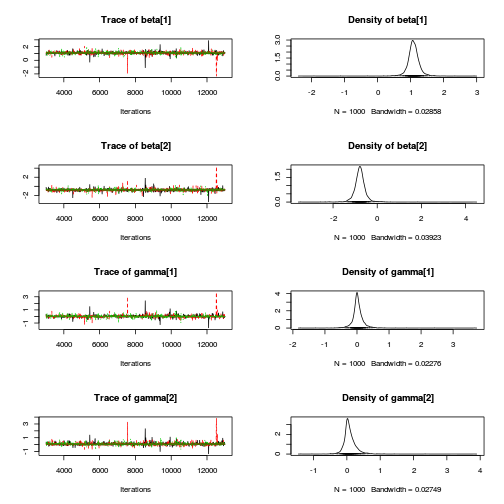
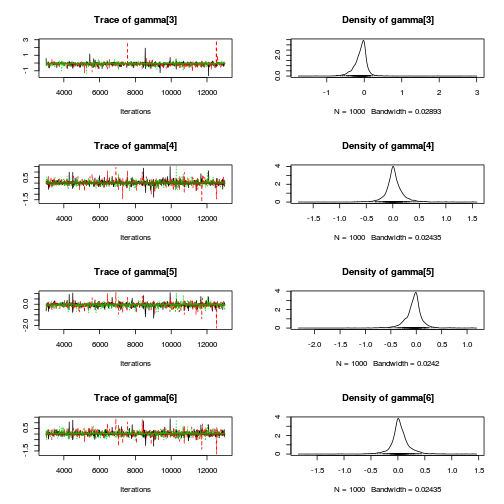
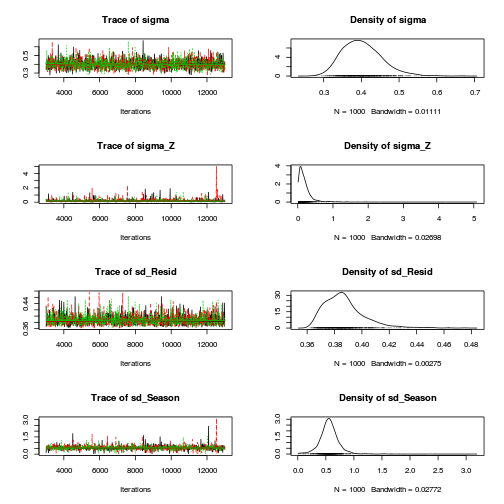
autocorr.diag(curdies.mcmc.d[, preds])
beta[1] beta[2] sigma sigma_Z Lag 0 1.000000 1.000000 1.000000 1.0000000 Lag 10 0.064133 0.081592 0.048868 0.2580142 Lag 50 0.047251 0.032703 0.003835 0.0200241 Lag 100 -0.004018 -0.007012 -0.005977 -0.0031332 Lag 500 -0.004367 0.001529 -0.002180 -0.0002983
raftery.diag(curdies.mcmc.d)
[[1]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[2]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[3]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
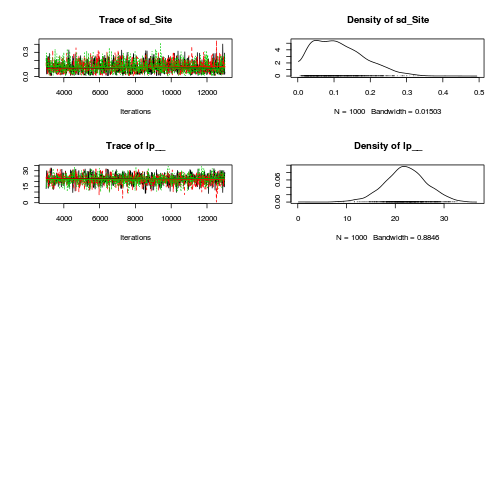
Hierarchical parameterization STAN codepreds <- c("beta[1]", "beta[2]", "sigma", "sigma_Z") plot(curdies.mcmc.e)
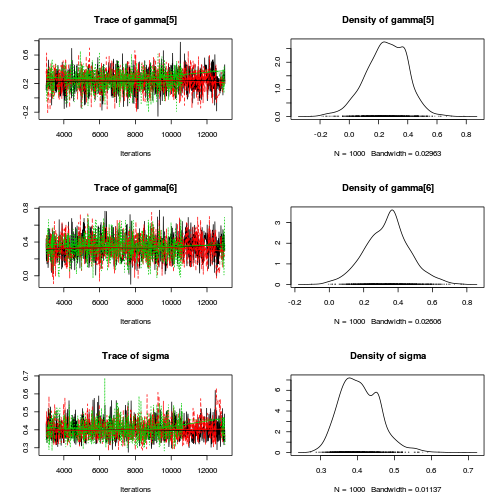
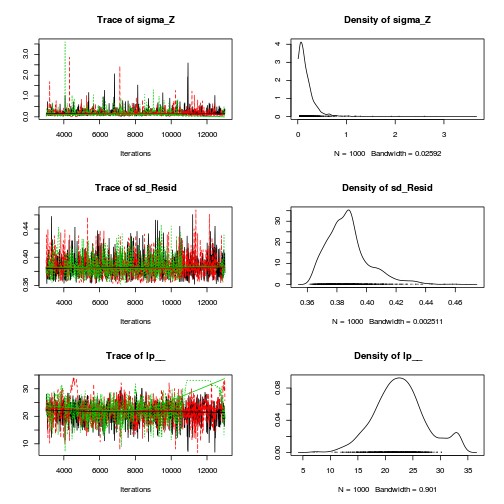
autocorr.diag(curdies.mcmc.e[, preds])
beta[1] beta[2] sigma sigma_Z Lag 0 1.0000000 1.000000 1.00000 1.000000 Lag 10 0.0006449 0.038216 0.21003 0.178918 Lag 50 0.0177504 0.011618 0.14116 0.045590 Lag 100 -0.0149410 -0.051226 0.11878 0.068740 Lag 500 0.0262724 0.008045 0.07296 0.002794
raftery.diag(curdies.mcmc.e)
[[1]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[2]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[3]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
- The variances in particular display substantial autocorrelation, perhaps it is worth exploring
a weekly informative Half-Cauchy prior (scale = 25) rather than a non-informative flat prior (as suggested by Gelman). It
might also be worth increasing the thinning rate from 10 to 100.
Show full effects parameterization JAGS code
modelString=" model { #Likelihood for (i in 1:n) { y[i]~dnorm(mu[i],tau) mu[i] <- alpha0 + alpha[SEASON[i]] + beta[SITE[i]] y.err[i] <- y[i]-mu[i] } #Priors alpha0 ~ dnorm(0, 1.0E-6) alpha[1] <- 0 for (i in 2:nSEASON) { alpha[i] ~ dnorm(0, 1.0E-6) #prior } for (i in 1:nSITE) { beta[i] ~ dnorm(0, tau.B) #prior } #Half-cauchy weekly informative prior (scale=25) tau <- pow(sigma,-2) sigma <- z/sqrt(chSq) z ~ dnorm(0, .0016)I(0,) chSq ~ dgamma(0.5, 0.5) tau.B <- pow(sigma.B,-2) sigma.B <- z.B/sqrt(chSq.B) z.B ~ dnorm(0, .0016)I(0,) chSq.B ~ dgamma(0.5, 0.5) #Finite-population standard deviations for (i in 1:nSEASON) { SD.Site[i] <- sd(beta[SiteOffset[i]:SiteOffset[i+1]-1]) } sd.Season <- sd(alpha) sd.Site <- mean(SD.Site) #sd(beta.site) sd.Resid <- sd(y.err) } " writeLines(modelString,con="../downloads/BUGSscripts/ws9.2bS1.2a.txt") # Generate a data list curdies$SEASON <- factor(curdies$SEASON, levels=c("WINTER","SUMMER")) sites<-ddply(curdies, ~SITE, catcolwise(unique)) offset <- c(match(levels(sites$SEASON),sites$SEASON), length(sites$SEASON)+1) offset
[1] 1 4 7
curdies$SITE <- factor(curdies$SITE) curdies.list <- with(curdies, list(y=S4DUGES, SITE=as.numeric(SITE), SEASON=as.numeric(SEASON), n=nrow(curdies), nSITE=length(levels(SITE)), nSEASON = length(levels(SEASON)), SiteOffset=offset ) ) # define parameters params <- c("alpha0","alpha","sigma","sigma.B","sd.Season","sd.Site","sd.Resid","SD.Site") burnInSteps = 6000 nChains = 3 numSavedSteps = 3000 thinSteps = 100 nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) library(R2jags) curdies.r2jags.a <- jags(data=curdies.list, inits=NULL, parameters.to.save=params, model.file="../downloads/BUGSscripts/ws9.2bS1.2a.txt", n.chains=nChains, n.iter=nIter, n.burnin=burnInSteps, n.thin=thinSteps )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 182 Initializing model
print(curdies.r2jags.a)
Inference for Bugs model at "../downloads/BUGSscripts/ws9.2bS1.2a.txt", fit using jags, 3 chains, each with 106000 iterations (first 6000 discarded), n.thin = 100 n.sims = 3000 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff SD.Site[1] 0.116 0.093 0.004 0.040 0.096 0.172 0.337 1.001 3000 SD.Site[2] 0.092 0.072 0.003 0.035 0.078 0.133 0.268 1.001 3000 alpha[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 alpha[2] -0.787 0.238 -1.252 -0.910 -0.786 -0.662 -0.317 1.003 3000 alpha0 1.091 0.167 0.767 1.000 1.092 1.178 1.423 1.002 3000 sd.Resid 0.388 0.015 0.366 0.378 0.386 0.395 0.426 1.001 3000 sd.Season 0.558 0.163 0.229 0.468 0.556 0.644 0.885 1.007 3000 sd.Site 0.104 0.071 0.005 0.046 0.094 0.150 0.262 1.001 3000 sigma 0.403 0.052 0.316 0.366 0.398 0.434 0.519 1.001 3000 sigma.B 0.173 0.170 0.006 0.064 0.130 0.224 0.618 1.001 3000 deviance 35.064 3.389 29.936 32.646 34.543 36.939 43.121 1.001 3000 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 5.7 and DIC = 40.8 DIC is an estimate of expected predictive error (lower deviance is better).curdies.mcmc.a <- as.mcmc(curdies.r2jags.a) preds <- c("alpha0", "alpha[2]", "sigma", "sigma.B") plot(curdies.mcmc.a)
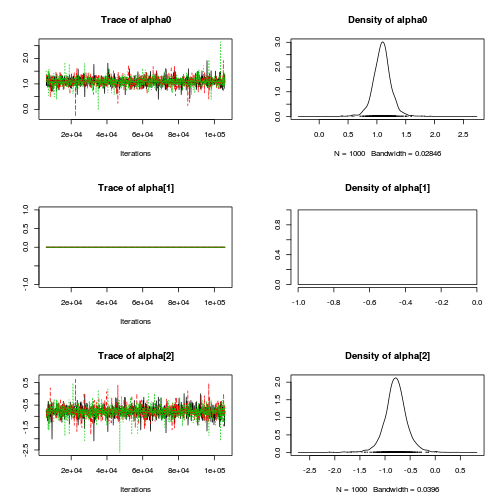
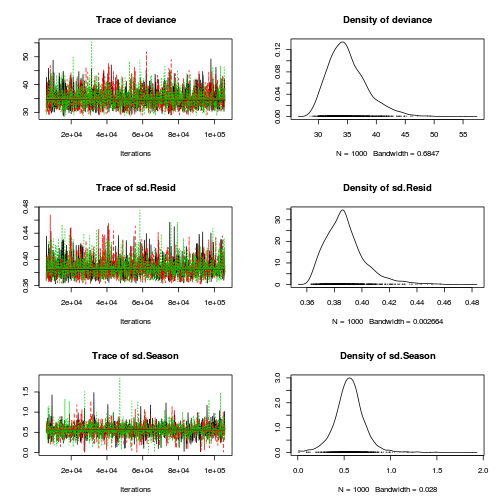
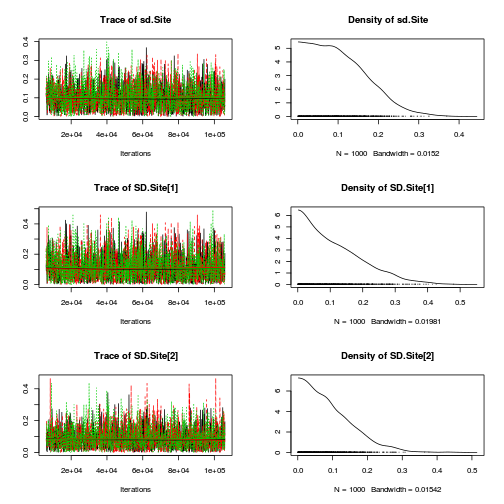
autocorr.diag(curdies.mcmc.a[, preds])
alpha0 alpha[2] sigma sigma.B Lag 0 1.000000 1.000000 1.000000 1.000000 Lag 100 -0.009808 0.007488 0.007509 0.011979 Lag 500 0.015313 0.006902 0.010719 0.004157 Lag 1000 -0.004642 -0.031708 0.019999 -0.035305 Lag 5000 -0.021117 0.001414 -0.007450 0.005227
raftery.diag(curdies.mcmc.a)
[[1]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[2]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[3]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
Show matrix parameterization JAGS codemodelString=" model { #Likelihood for (i in 1:n) { y[i]~dnorm(mu[i],tau) mu[i] <- inprod(alpha[],X[i,]) + beta[SITE[i]] y.err[i] <- y[i] - mu[i] } #Priors alpha ~ dmnorm(a0, A0) for (i in 1:nSITE) { beta[i] ~ dnorm(0, tau.B) #prior } #Half-cauchy weekly informative prior (scale=25) tau <- pow(sigma,-2) sigma <- z/sqrt(chSq) z ~ dnorm(0, .0016)I(0,) chSq ~ dgamma(0.5, 0.5) tau.B <- pow(sigma.B,-2) sigma.B <- z.B/sqrt(chSq.B) z.B ~ dnorm(0, .0016)I(0,) chSq.B ~ dgamma(0.5, 0.5) # Finite-population standard deviations for (i in 1:nSEASON) { SD.Site[i] <- sd(beta[SiteOffset[i]:SiteOffset[i+1]-1]) } seasons[1] <- alpha[1] for (i in 2:nSEASON) { seasons[i] <- alpha[1]+alpha[i] } sd.Season <- sd(seasons) sd.Site <- mean(SD.Site) sd.Resid <- sd(y.err) } " writeLines(modelString,con="../downloads/BUGSscripts/ws9.2bS1.2b.txt") # Generate a data list curdies$SEASON <- factor(curdies$SEASON, levels=c("WINTER","SUMMER")) sites<-ddply(curdies, ~SITE, catcolwise(unique)) offset <- c(match(levels(sites$SEASON),sites$SEASON), length(sites$SEASON)+1) curdies$SITE <- factor(curdies$SITE) Xmat <- model.matrix(~SEASON, data=curdies) curdies.list <- with(curdies, list(y=S4DUGES, SITE=as.numeric(SITE), X=Xmat, n=nrow(curdies), nSEASON=length(levels(SEASON)), nSITE=length(levels(SITE)), a0=rep(0, ncol(Xmat)), A0=diag(0, ncol(Xmat)), SiteOffset=offset ) ) # define parameters params <- c("alpha","sigma","sigma.B","sd.Season","sd.Site","sd.Resid","SD.Site") burnInSteps = 6000 nChains = 3 numSavedSteps = 3000 thinSteps = 100 nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) library(R2jags) curdies.r2jags.b <- jags(data=curdies.list, inits=NULL, parameters.to.save=params, model.file="../downloads/BUGSscripts/ws9.2bS1.2b.txt", n.chains=nChains, n.iter=nIter, n.burnin=burnInSteps, n.thin=thinSteps )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 264 Initializing model
print(curdies.r2jags.b)
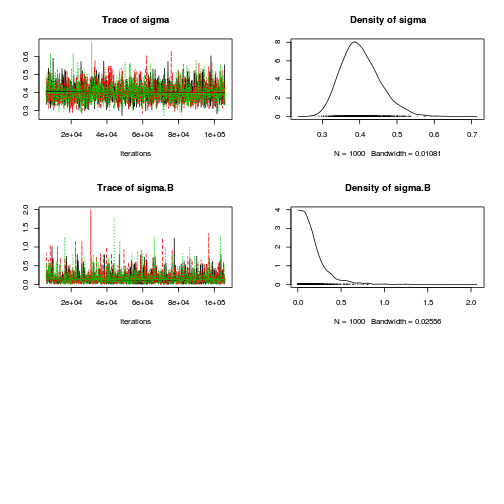
Inference for Bugs model at "../downloads/BUGSscripts/ws9.2bS1.2b.txt", fit using jags, 3 chains, each with 106000 iterations (first 6000 discarded), n.thin = 100 n.sims = 3000 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff SD.Site[1] 0.118 0.092 0.004 0.043 0.097 0.175 0.335 1.001 3000 SD.Site[2] 0.093 0.072 0.004 0.038 0.078 0.132 0.264 1.003 1500 alpha[1] 1.092 0.199 0.744 1.002 1.093 1.182 1.407 1.027 3000 alpha[2] -0.789 0.272 -1.230 -0.909 -0.789 -0.663 -0.309 1.022 3000 sd.Resid 0.388 0.015 0.366 0.378 0.386 0.394 0.425 1.004 650 sd.Season 0.561 0.182 0.238 0.470 0.558 0.643 0.870 1.001 3000 sd.Site 0.105 0.071 0.005 0.049 0.095 0.149 0.273 1.001 3000 sigma 0.401 0.052 0.313 0.364 0.397 0.433 0.513 1.003 760 sigma.B 0.181 0.190 0.007 0.066 0.134 0.235 0.666 1.001 3000 deviance 35.065 3.456 29.843 32.650 34.563 36.879 43.448 1.003 970 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 6.0 and DIC = 41.0 DIC is an estimate of expected predictive error (lower deviance is better).curdies.mcmc.b <- as.mcmc(curdies.r2jags.b) preds <- c("alpha[1]", "alpha[2]", "sigma", "sigma.B") plot(curdies.mcmc.b)
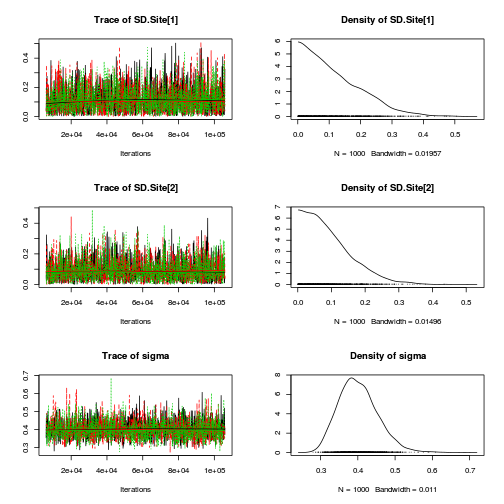
autocorr.diag(curdies.mcmc.b[, preds])
alpha[1] alpha[2] sigma sigma.B Lag 0 1.000000 1.000000 1.00000 1.00000 Lag 100 -0.014878 -0.002648 -0.03852 0.08885 Lag 500 -0.006045 0.021407 0.01152 0.01435 Lag 1000 0.017723 0.014398 0.01089 0.01031 Lag 5000 -0.024461 -0.013764 -0.01254 -0.01210
raftery.diag(curdies.mcmc.b)
[[1]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[2]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[3]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
Hierarchical parameterization JAGS codemodelString= writeLines(modelString,con="../downloads/BUGSscripts/ws9.2bS1.2c.txt") # Generate a data list curdies$SEASON <- factor(curdies$SEASON, levels=c("WINTER","SUMMER")) sites<-ddply(curdies, ~SITE, catcolwise(unique)) offset <- c(match(levels(sites$SEASON),sites$SEASON), length(sites$SEASON)+1) curdies$SITE <- factor(curdies$SITE) Xmat <- model.matrix(~SEASON, data=ddply(curdies, ~SITE, catcolwise(unique))) curdies.list <- with(curdies, list(y=S4DUGES, SITE=as.numeric(SITE), X=Xmat, nX=ncol(Xmat), n=nrow(curdies), nSITE=length(levels(SITE)), nSEASON=length(levels(SEASON)), SiteOffset=offset ) ) # define parameters params <- c("beta","sigma","sigma.site","sd.Season","sd.Site","sd.Resid") burnInSteps = 6000 nChains = 3 numSavedSteps = 3000 thinSteps = 100 nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) library(R2jags) curdies.r2jags.c <- jags(data=curdies.list, inits=NULL, parameters.to.save=params, model.file="../downloads/BUGSscripts/ws9.2bS1.2c.txt", n.chains=nChains, n.iter=nIter, n.burnin=burnInSteps, n.thin=thinSteps )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 162 Initializing model
print(curdies.r2jags.c)
Inference for Bugs model at "../downloads/BUGSscripts/ws9.2bS1.2c.txt", fit using jags, 3 chains, each with 106000 iterations (first 6000 discarded), n.thin = 100 n.sims = 3000 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff beta[1] 1.094 0.189 0.773 1.006 1.094 1.180 1.436 1.008 3000 beta[2] -0.793 0.269 -1.269 -0.914 -0.790 -0.663 -0.331 1.002 3000 sd.Resid 0.387 0.015 0.366 0.377 0.385 0.394 0.424 1.001 3000 sd.Season 0.564 0.181 0.239 0.470 0.559 0.647 0.898 1.004 3000 sd.Site 0.105 0.070 0.005 0.049 0.096 0.148 0.268 1.002 1900 sigma 0.403 0.053 0.315 0.365 0.398 0.435 0.522 1.001 3000 sigma.site 0.178 0.194 0.008 0.063 0.130 0.229 0.628 1.002 1600 deviance 35.009 3.489 29.758 32.590 34.487 36.748 43.487 1.001 3000 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 6.1 and DIC = 41.1 DIC is an estimate of expected predictive error (lower deviance is better).curdies.mcmc.c <- as.mcmc(curdies.r2jags.c) preds <- c("beta[1]", "beta[2]", "sigma", "sigma.site") plot(curdies.mcmc.c)
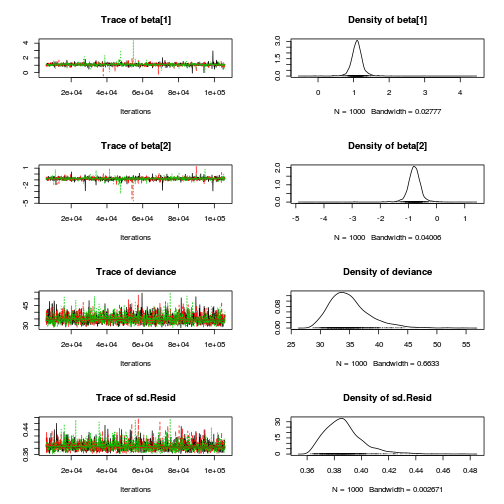
autocorr.diag(curdies.mcmc.c[, preds])
beta[1] beta[2] sigma sigma.site Lag 0 1.000000 1.0000000 1.00000 1.000000 Lag 100 0.009198 0.0321138 -0.01952 0.066553 Lag 500 -0.021866 0.0169660 0.01633 -0.020410 Lag 1000 -0.014282 -0.0008585 -0.01030 -0.016491 Lag 5000 -0.020213 0.0011308 0.01814 0.008906
raftery.diag(curdies.mcmc.c)
[[1]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[2]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[3]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
Matrix parameterization STAN coderstanString=" data{ int n; int nZ; int nX; vector [n] y; matrix [n,nX] X; matrix [n,nZ] Z; vector [nX] a0; matrix [nX,nX] A0; } parameters{ vector [nX] beta; real<lower=0> sigma; vector [nZ] gamma; real<lower=0> sigma_Z; } transformed parameters { vector [n] mu; mu <- X*beta + Z*gamma; } model{ // Priors beta ~ multi_normal(a0,A0); gamma ~ normal( 0 , sigma_Z ); sigma_Z ~ cauchy(0,25); sigma ~ cauchy(0,25); y ~ normal( mu , sigma ); } generated quantities { vector [n] y_err; real<lower=0> sd_Resid; y_err <- y - mu; sd_Resid <- sd(y_err); } " # Generate a data list curdies$SITE <- factor(curdies$SITE) Xmat <- model.matrix(~SEASON, data=curdies) Zmat <- model.matrix(~-1+SITE, data=curdies) curdies.list <- with(curdies, list(y=S4DUGES, Z=Zmat, X=Xmat, nX=ncol(Xmat), nZ=ncol(Zmat), n=nrow(curdies), a0=rep(0,ncol(Xmat)), A0=diag(100000,ncol(Xmat)) ) ) # define parameters burnInSteps = 6000 nChains = 3 numSavedSteps = 3000 thinSteps = 100 nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) library(rstan) curdies.rstan.a <- stan(data=curdies.list, model_code=rstanString, pars=c('beta','gamma','sigma','sigma_Z', 'sd_Resid'), chains=nChains, iter=nIter, warmup=burnInSteps, thin=thinSteps, save_dso=TRUE )
TRANSLATING MODEL 'rstanString' FROM Stan CODE TO C++ CODE NOW. COMPILING THE C++ CODE FOR MODEL 'rstanString' NOW. SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 1). Iteration: 1 / 106000 [ 0%] (Warmup) Iteration: 6001 / 106000 [ 5%] (Sampling) Iteration: 16600 / 106000 [ 15%] (Sampling) Iteration: 27200 / 106000 [ 25%] (Sampling) Iteration: 37800 / 106000 [ 35%] (Sampling) Iteration: 48400 / 106000 [ 45%] (Sampling) Iteration: 59000 / 106000 [ 55%] (Sampling) Iteration: 69600 / 106000 [ 65%] (Sampling) Iteration: 80200 / 106000 [ 75%] (Sampling) Iteration: 90800 / 106000 [ 85%] (Sampling) Iteration: 101400 / 106000 [ 95%] (Sampling) Iteration: 106000 / 106000 [100%] (Sampling) # Elapsed Time: 0.65 seconds (Warm-up) # 13.74 seconds (Sampling) # 14.39 seconds (Total) SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 2). Iteration: 1 / 106000 [ 0%] (Warmup) Iteration: 6001 / 106000 [ 5%] (Sampling) Iteration: 16600 / 106000 [ 15%] (Sampling) Iteration: 27200 / 106000 [ 25%] (Sampling) Iteration: 37800 / 106000 [ 35%] (Sampling) Iteration: 48400 / 106000 [ 45%] (Sampling) Iteration: 59000 / 106000 [ 55%] (Sampling) Iteration: 69600 / 106000 [ 65%] (Sampling) Iteration: 80200 / 106000 [ 75%] (Sampling) Iteration: 90800 / 106000 [ 85%] (Sampling) Iteration: 101400 / 106000 [ 95%] (Sampling) Iteration: 106000 / 106000 [100%] (Sampling) # Elapsed Time: 0.64 seconds (Warm-up) # 11.84 seconds (Sampling) # 12.48 seconds (Total) SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 3). Iteration: 1 / 106000 [ 0%] (Warmup) Iteration: 6001 / 106000 [ 5%] (Sampling) Iteration: 16600 / 106000 [ 15%] (Sampling) Iteration: 27200 / 106000 [ 25%] (Sampling) Iteration: 37800 / 106000 [ 35%] (Sampling) Iteration: 48400 / 106000 [ 45%] (Sampling) Iteration: 59000 / 106000 [ 55%] (Sampling) Iteration: 69600 / 106000 [ 65%] (Sampling) Iteration: 80200 / 106000 [ 75%] (Sampling) Iteration: 90800 / 106000 [ 85%] (Sampling) Iteration: 101400 / 106000 [ 95%] (Sampling) Iteration: 106000 / 106000 [100%] (Sampling) # Elapsed Time: 0.72 seconds (Warm-up) # 12.08 seconds (Sampling) # 12.8 seconds (Total)
print(curdies.rstan.a)
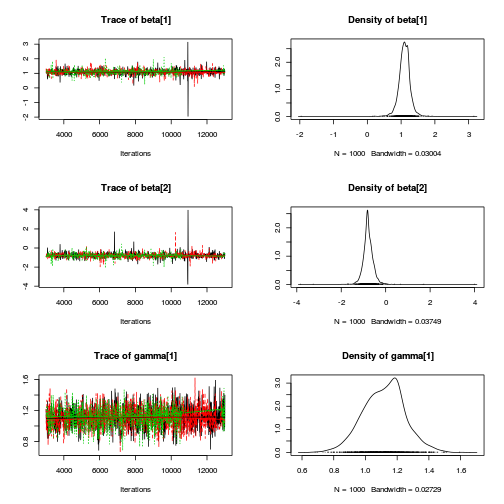
Inference for Stan model: rstanString. 3 chains, each with iter=106000; warmup=6000; thin=100; post-warmup draws per chain=1000, total post-warmup draws=3000. mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat beta[1] 1.09 0.00 0.18 0.76 1.00 1.10 1.19 1.43 2991 1 beta[2] -0.79 0.00 0.25 -1.26 -0.91 -0.79 -0.66 -0.31 2941 1 gamma[1] 0.02 0.00 0.17 -0.34 -0.05 0.01 0.08 0.38 2935 1 gamma[2] 0.08 0.00 0.18 -0.23 -0.01 0.05 0.15 0.50 2804 1 gamma[3] -0.10 0.00 0.19 -0.53 -0.18 -0.06 0.00 0.18 3000 1 gamma[4] 0.02 0.00 0.17 -0.31 -0.05 0.01 0.08 0.39 3000 1 gamma[5] -0.06 0.00 0.18 -0.47 -0.12 -0.03 0.02 0.26 2812 1 gamma[6] 0.04 0.00 0.17 -0.30 -0.04 0.02 0.10 0.42 3000 1 sigma 0.40 0.00 0.05 0.32 0.36 0.40 0.43 0.51 2658 1 sigma_Z 0.19 0.00 0.18 0.02 0.07 0.14 0.24 0.65 2700 1 sd_Resid 0.39 0.00 0.01 0.37 0.38 0.39 0.39 0.42 2878 1 lp__ 22.54 0.09 4.47 13.90 19.66 22.42 25.43 31.65 2635 1 Samples were drawn using NUTS(diag_e) at Wed Jan 28 13:26:47 2015. For each parameter, n_eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence, Rhat=1).curdies.mcmc.d <- rstan:::as.mcmc.list.stanfit(curdies.rstan.a) curdies.mcmc.df.d <- as.data.frame(extract(curdies.rstan.a)) preds <- c("beta[1]", "beta[2]", "sigma", "sigma_Z") plot(curdies.mcmc.d)
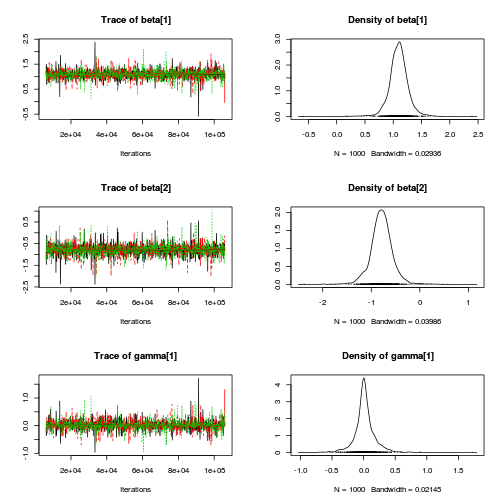
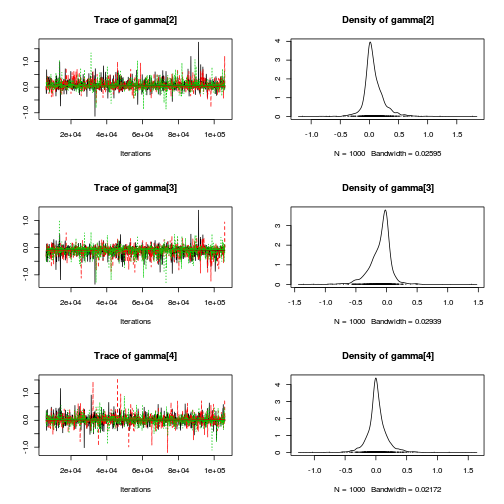
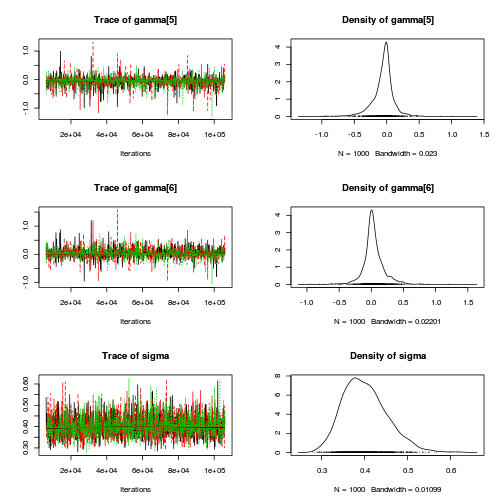
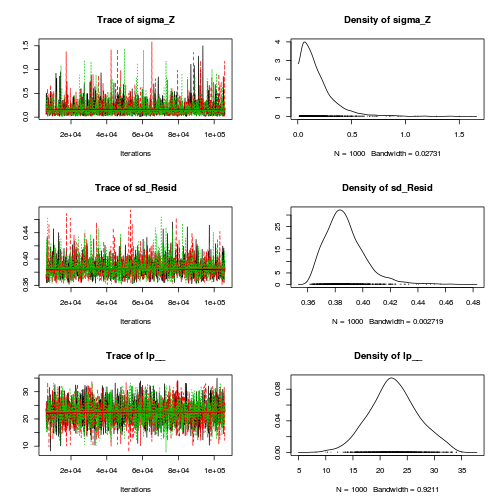
autocorr.diag(curdies.mcmc.d[, preds])
beta[1] beta[2] sigma sigma_Z Lag 0 1.00000 1.000000 1.000000 1.00000 Lag 100 0.00166 0.012112 0.022410 0.01175 Lag 500 -0.01420 -0.005727 0.018801 0.00282 Lag 1000 0.01339 0.016978 0.004863 -0.01002 Lag 5000 0.00539 -0.009073 0.025374 -0.03148
raftery.diag(curdies.mcmc.d)
[[1]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[2]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[3]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
#Finite-population standard deviations ## Site curdies$SEASON <- factor(curdies$SEASON, levels=c("WINTER","SUMMER")) sites<-ddply(curdies, ~SITE, catcolwise(unique))
Error: could not find function "ddply"
offset <- c(match(levels(sites$SEASON),sites$SEASON), length(sites$SEASON)+1) SD.site <- NULL sd.site <- curdies.mcmc.df.d[,3:8] for (i in 1:length(levels(curdies$SEASON))) { SD.site <- c(SD.site,apply(sd.site[,offset[i]:(offset[i+1]-1)],1,sd)) } data.frame(mean=mean(SD.site), median=median(SD.site), HPDinterval(as.mcmc(SD.site)), HPDinterval(as.mcmc(SD.site),p=0.68))
mean median lower upper lower.1 upper.1 var1 0.1097 0.08922 0.0009367 0.2719 0.007624 0.1381
#Season seasons <- NULL seasons <- cbind(curdies.mcmc.df.d[,'beta.1'], curdies.mcmc.df.d[,'beta.1']+curdies.mcmc.df.d[,'beta.2']) sd.season <- apply(seasons,1,sd) library(coda) data.frame(mean=mean(sd.season), median=median(sd.season), HPDinterval(as.mcmc(sd.season)), HPDinterval(as.mcmc(sd.season),p=0.68))
mean median lower upper lower.1 upper.1 var1 0.5605 0.5582 0.2427 0.901 0.4325 0.7008
Hierarchical parameterization STAN coderstanString=" data{ int n; int nZ; int nX; vector [n] y; matrix [nZ,nX] X; matrix [n,nZ] Z; } parameters{ vector [nX] beta; real<lower=0> sigma; vector [nZ] gamma; real<lower=0> sigma_Z; } transformed parameters { vector [n] mu_Z; vector [nZ] mu; mu_Z <- Z*gamma; mu <- X*beta; } model{ // Priors beta ~ normal(0, 10000); gamma ~ normal( 0 , 1000); sigma_Z ~ uniform( 0 , 100 ); sigma ~ uniform( 0 , 100 ); y ~ normal(mu_Z, sigma); gamma ~ normal( mu , sigma_Z ); } generated quantities { vector [n] y_err; real<lower=0> sd_Resid; y_err <- y - mu_Z; sd_Resid <- sd(y_err); } " # Generate a data list curdies$SITE <- factor(curdies$SITE) Xmat <- model.matrix(~SEASON, data=ddply(curdies, ~SITE, catcolwise(unique))) Zmat <- model.matrix(~-1+SITE, data=curdies) curdies.list <- with(curdies, list(y=S4DUGES, Z=Zmat, X=Xmat, nX=ncol(Xmat), nZ=ncol(Zmat), n=nrow(curdies) ) ) curdies.list
$y [1] 0.8971 1.5713 1.0700 1.1461 1.0991 1.0141 1.0040 2.0061 1.0396 1.0060 1.1909 1.3845 0.9111 [14] 1.0508 1.0272 0.9003 0.6074 0.7104 0.0000 0.0000 0.9849 0.0000 0.0000 1.1200 1.0828 0.0000 [27] 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.6038 0.0000 0.9007 0.7823 $Z SITE1 SITE2 SITE3 SITE4 SITE5 SITE6 1 1 0 0 0 0 0 2 1 0 0 0 0 0 3 1 0 0 0 0 0 4 1 0 0 0 0 0 5 1 0 0 0 0 0 6 1 0 0 0 0 0 7 0 1 0 0 0 0 8 0 1 0 0 0 0 9 0 1 0 0 0 0 10 0 1 0 0 0 0 11 0 1 0 0 0 0 12 0 1 0 0 0 0 13 0 0 1 0 0 0 14 0 0 1 0 0 0 15 0 0 1 0 0 0 16 0 0 1 0 0 0 17 0 0 1 0 0 0 18 0 0 1 0 0 0 19 0 0 0 1 0 0 20 0 0 0 1 0 0 21 0 0 0 1 0 0 22 0 0 0 1 0 0 23 0 0 0 1 0 0 24 0 0 0 1 0 0 25 0 0 0 0 1 0 26 0 0 0 0 1 0 27 0 0 0 0 1 0 28 0 0 0 0 1 0 29 0 0 0 0 1 0 30 0 0 0 0 1 0 31 0 0 0 0 0 1 32 0 0 0 0 0 1 33 0 0 0 0 0 1 34 0 0 0 0 0 1 35 0 0 0 0 0 1 36 0 0 0 0 0 1 attr(,"assign") [1] 1 1 1 1 1 1 attr(,"contrasts") attr(,"contrasts")$SITE [1] "contr.treatment" $X (Intercept) SEASONSUMMER 1 1 0 2 1 0 3 1 0 4 1 1 5 1 1 6 1 1 attr(,"assign") [1] 0 1 attr(,"contrasts") attr(,"contrasts")$SEASON [1] "contr.treatment" $nX [1] 2 $nZ [1] 6 $n [1] 36
# define parameters burnInSteps = 6000 nChains = 3 numSavedSteps = 3000 thinSteps = 100 nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) library(rstan) curdies.rstan.b <- stan(data=curdies.list, model_code=rstanString, pars=c('beta','gamma','sigma','sigma_Z','sd_Resid'), chains=nChains, iter=nIter, warmup=burnInSteps, thin=thinSteps, save_dso=TRUE )
TRANSLATING MODEL 'rstanString' FROM Stan CODE TO C++ CODE NOW. COMPILING THE C++ CODE FOR MODEL 'rstanString' NOW. SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 1). Iteration: 1 / 106000 [ 0%] (Warmup) Iteration: 6001 / 106000 [ 5%] (Sampling) Iteration: 16600 / 106000 [ 15%] (Sampling) Iteration: 27200 / 106000 [ 25%] (Sampling) Iteration: 37800 / 106000 [ 35%] (Sampling) Iteration: 48400 / 106000 [ 45%] (Sampling) Iteration: 59000 / 106000 [ 55%] (Sampling) Iteration: 69600 / 106000 [ 65%] (Sampling) Iteration: 80200 / 106000 [ 75%] (Sampling) Iteration: 90800 / 106000 [ 85%] (Sampling) Iteration: 101400 / 106000 [ 95%] (Sampling) Iteration: 106000 / 106000 [100%] (Sampling) # Elapsed Time: 0.65 seconds (Warm-up) # 16.77 seconds (Sampling) # 17.42 seconds (Total) SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 2). Iteration: 1 / 106000 [ 0%] (Warmup) Iteration: 6001 / 106000 [ 5%] (Sampling) Iteration: 16600 / 106000 [ 15%] (Sampling) Iteration: 27200 / 106000 [ 25%] (Sampling) Iteration: 37800 / 106000 [ 35%] (Sampling) Iteration: 48400 / 106000 [ 45%] (Sampling) Iteration: 59000 / 106000 [ 55%] (Sampling) Iteration: 69600 / 106000 [ 65%] (Sampling) Iteration: 80200 / 106000 [ 75%] (Sampling) Iteration: 90800 / 106000 [ 85%] (Sampling) Iteration: 101400 / 106000 [ 95%] (Sampling) Iteration: 106000 / 106000 [100%] (Sampling) # Elapsed Time: 0.73 seconds (Warm-up) # 22.38 seconds (Sampling) # 23.11 seconds (Total) SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 3). Iteration: 1 / 106000 [ 0%] (Warmup) Iteration: 6001 / 106000 [ 5%] (Sampling) Iteration: 16600 / 106000 [ 15%] (Sampling) Iteration: 27200 / 106000 [ 25%] (Sampling) Iteration: 37800 / 106000 [ 35%] (Sampling) Iteration: 48400 / 106000 [ 45%] (Sampling) Iteration: 59000 / 106000 [ 55%] (Sampling) Iteration: 69600 / 106000 [ 65%] (Sampling) Iteration: 80200 / 106000 [ 75%] (Sampling) Iteration: 90800 / 106000 [ 85%] (Sampling) Iteration: 101400 / 106000 [ 95%] (Sampling) Iteration: 106000 / 106000 [100%] (Sampling) # Elapsed Time: 0.59 seconds (Warm-up) # 7.32 seconds (Sampling) # 7.91 seconds (Total)
print(curdies.rstan.b)
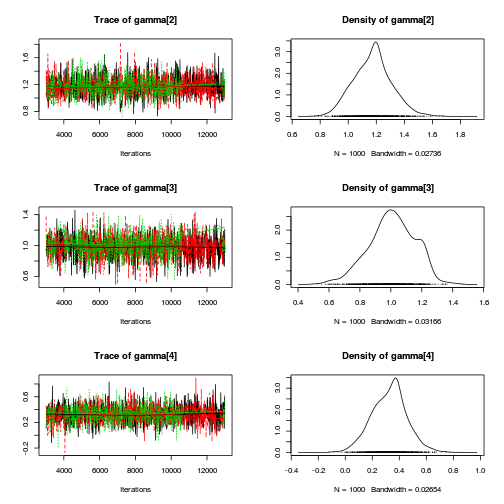
Inference for Stan model: rstanString. 3 chains, each with iter=106000; warmup=6000; thin=100; post-warmup draws per chain=1000, total post-warmup draws=3000. mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat beta[1] 1.10 0.00 0.17 0.78 1.01 1.10 1.19 1.44 1586 1.00 beta[2] -0.80 0.01 0.29 -1.28 -0.91 -0.80 -0.67 -0.35 3000 1.00 gamma[1] 1.12 0.01 0.13 0.87 1.03 1.12 1.20 1.37 594 1.01 gamma[2] 1.17 0.00 0.14 0.91 1.08 1.17 1.26 1.46 1908 1.00 gamma[3] 1.00 0.01 0.15 0.68 0.91 1.01 1.11 1.32 216 1.01 gamma[4] 0.33 0.00 0.13 0.06 0.24 0.33 0.42 0.59 1197 1.00 gamma[5] 0.26 0.01 0.14 -0.03 0.17 0.26 0.35 0.50 445 1.01 gamma[6] 0.34 0.00 0.13 0.10 0.25 0.34 0.42 0.61 1305 1.00 sigma 0.40 0.00 0.05 0.31 0.37 0.40 0.43 0.52 2051 1.00 sigma_Z 0.18 0.00 0.21 0.02 0.06 0.12 0.23 0.65 2315 1.00 sd_Resid 0.39 0.00 0.01 0.37 0.38 0.39 0.39 0.42 3000 1.00 lp__ 22.74 0.19 4.62 13.47 19.87 22.76 25.57 31.85 599 1.00 Samples were drawn using NUTS(diag_e) at Wed Jan 28 13:32:21 2015. For each parameter, n_eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence, Rhat=1).curdies.mcmc.e <- rstan:::as.mcmc.list.stanfit(curdies.rstan.b) curdies.mcmc.df.e <- as.data.frame(extract(curdies.rstan.b)) preds <- c("beta[1]", "beta[2]", "sigma", "sigma_Z") plot(curdies.mcmc.e)
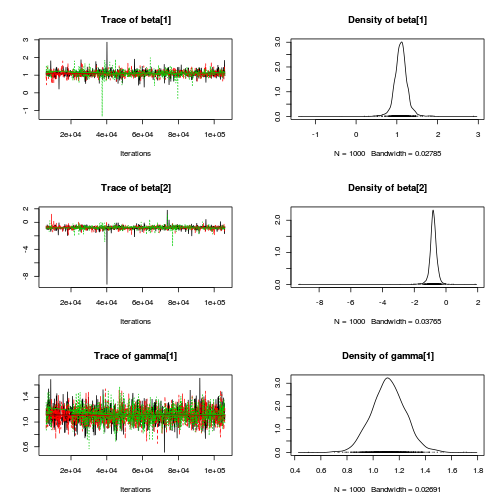
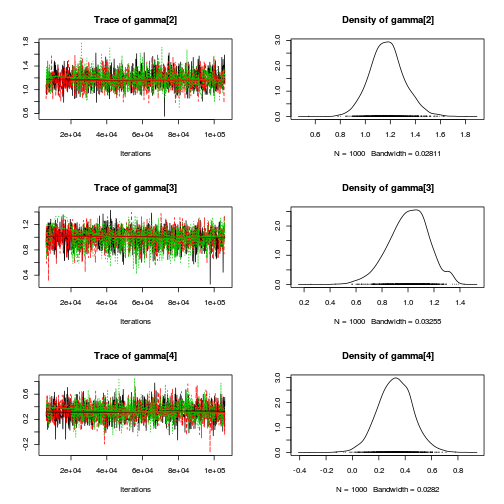
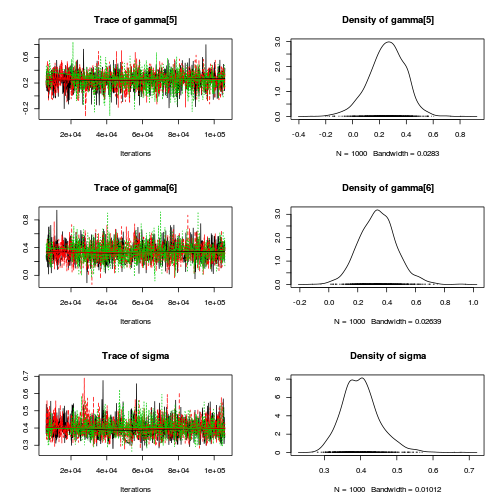
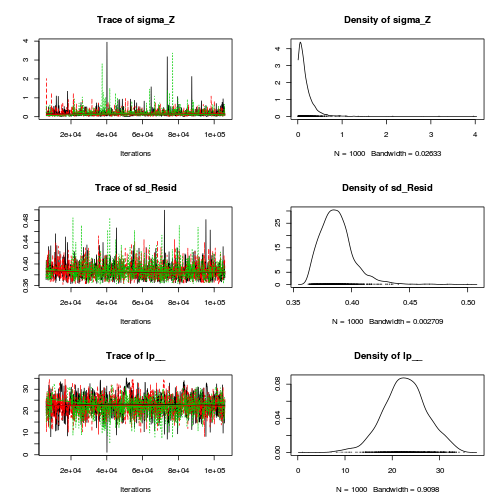
autocorr.diag(curdies.mcmc.e[, preds])
beta[1] beta[2] sigma sigma_Z Lag 0 1.000000 1.0000000 1.000000 1.00000 Lag 100 0.002777 -0.0232722 0.041328 0.06887 Lag 500 0.016810 -0.0005176 0.045240 0.02156 Lag 1000 0.016108 -0.0021485 -0.006067 0.02749 Lag 5000 -0.001203 -0.0261897 -0.005519 0.01899
raftery.diag(curdies.mcmc.e)
[[1]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[2]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[3]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
#Finite-population standard deviations ## Seasons seasons <- NULL seasons <- cbind(curdies.mcmc.df.e[,'beta.1'], curdies.mcmc.df.e[,'beta.1']+curdies.mcmc.df.e[,'beta.2']) sd.season <- apply(seasons,1,sd) library(coda) data.frame(mean=mean(sd.season), median=median(sd.season), HPDinterval(as.mcmc(sd.season)), HPDinterval(as.mcmc(sd.season),p=0.68))
mean median lower upper lower.1 upper.1 var1 0.5669 0.5645 0.239 0.8725 0.4288 0.6815
## Sites SD.site <- NULL sd.site <- curdies.mcmc.df.e[,3:8] for (i in 1:length(levels(curdies$SEASON))) { SD.site <- c(SD.site,apply(sd.site[,offset[i]:(offset[i+1]-1)],1,sd)) } data.frame(mean=mean(SD.site), median=median(SD.site), HPDinterval(as.mcmc(SD.site)), HPDinterval(as.mcmc(SD.site),p=0.68))
mean median lower upper lower.1 upper.1 var1 0.1077 0.08733 0.002412 0.2759 0.004596 0.1326
- Explore the parameter estimates
Full effects parameterization JAGS code
library(R2jags) print(curdies.r2jags.a)
Inference for Bugs model at "../downloads/BUGSscripts/ws9.2bS1.2a.txt", fit using jags, 3 chains, each with 106000 iterations (first 6000 discarded), n.thin = 100 n.sims = 3000 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff SD.Site[1] 0.116 0.093 0.004 0.040 0.096 0.172 0.337 1.001 3000 SD.Site[2] 0.092 0.072 0.003 0.035 0.078 0.133 0.268 1.001 3000 alpha[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 alpha[2] -0.787 0.238 -1.252 -0.910 -0.786 -0.662 -0.317 1.003 3000 alpha0 1.091 0.167 0.767 1.000 1.092 1.178 1.423 1.002 3000 sd.Resid 0.388 0.015 0.366 0.378 0.386 0.395 0.426 1.001 3000 sd.Season 0.558 0.163 0.229 0.468 0.556 0.644 0.885 1.007 3000 sd.Site 0.104 0.071 0.005 0.046 0.094 0.150 0.262 1.001 3000 sigma 0.403 0.052 0.316 0.366 0.398 0.434 0.519 1.001 3000 sigma.B 0.173 0.170 0.006 0.064 0.130 0.224 0.618 1.001 3000 deviance 35.064 3.389 29.936 32.646 34.543 36.939 43.121 1.001 3000 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 5.7 and DIC = 40.8 DIC is an estimate of expected predictive error (lower deviance is better).Full effects parameterization JAGS codeprint(curdies.r2jags.b)
Inference for Bugs model at "../downloads/BUGSscripts/ws9.2bS1.2b.txt", fit using jags, 3 chains, each with 106000 iterations (first 6000 discarded), n.thin = 100 n.sims = 3000 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff SD.Site[1] 0.118 0.092 0.004 0.043 0.097 0.175 0.335 1.001 3000 SD.Site[2] 0.093 0.072 0.004 0.038 0.078 0.132 0.264 1.003 1500 alpha[1] 1.092 0.199 0.744 1.002 1.093 1.182 1.407 1.027 3000 alpha[2] -0.789 0.272 -1.230 -0.909 -0.789 -0.663 -0.309 1.022 3000 sd.Resid 0.388 0.015 0.366 0.378 0.386 0.394 0.425 1.004 650 sd.Season 0.561 0.182 0.238 0.470 0.558 0.643 0.870 1.001 3000 sd.Site 0.105 0.071 0.005 0.049 0.095 0.149 0.273 1.001 3000 sigma 0.401 0.052 0.313 0.364 0.397 0.433 0.513 1.003 760 sigma.B 0.181 0.190 0.007 0.066 0.134 0.235 0.666 1.001 3000 deviance 35.065 3.456 29.843 32.650 34.563 36.879 43.448 1.003 970 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 6.0 and DIC = 41.0 DIC is an estimate of expected predictive error (lower deviance is better).Hierarchical parameterization JAGS codeprint(curdies.r2jags.c)
Inference for Bugs model at "../downloads/BUGSscripts/ws9.2bS1.2c.txt", fit using jags, 3 chains, each with 106000 iterations (first 6000 discarded), n.thin = 100 n.sims = 3000 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff beta[1] 1.094 0.189 0.773 1.006 1.094 1.180 1.436 1.008 3000 beta[2] -0.793 0.269 -1.269 -0.914 -0.790 -0.663 -0.331 1.002 3000 sd.Resid 0.387 0.015 0.366 0.377 0.385 0.394 0.424 1.001 3000 sd.Season 0.564 0.181 0.239 0.470 0.559 0.647 0.898 1.004 3000 sd.Site 0.105 0.070 0.005 0.049 0.096 0.148 0.268 1.002 1900 sigma 0.403 0.053 0.315 0.365 0.398 0.435 0.522 1.001 3000 sigma.site 0.178 0.194 0.008 0.063 0.130 0.229 0.628 1.002 1600 deviance 35.009 3.489 29.758 32.590 34.487 36.748 43.487 1.001 3000 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 6.1 and DIC = 41.1 DIC is an estimate of expected predictive error (lower deviance is better).Full matrix parameterization STAN codeprint(curdies.rstan.a)
Inference for Stan model: rstanString. 3 chains, each with iter=106000; warmup=6000; thin=100; post-warmup draws per chain=1000, total post-warmup draws=3000. mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat beta[1] 1.09 0.00 0.18 0.76 1.00 1.10 1.19 1.43 2991 1 beta[2] -0.79 0.00 0.25 -1.26 -0.91 -0.79 -0.66 -0.31 2941 1 gamma[1] 0.02 0.00 0.17 -0.34 -0.05 0.01 0.08 0.38 2935 1 gamma[2] 0.08 0.00 0.18 -0.23 -0.01 0.05 0.15 0.50 2804 1 gamma[3] -0.10 0.00 0.19 -0.53 -0.18 -0.06 0.00 0.18 3000 1 gamma[4] 0.02 0.00 0.17 -0.31 -0.05 0.01 0.08 0.39 3000 1 gamma[5] -0.06 0.00 0.18 -0.47 -0.12 -0.03 0.02 0.26 2812 1 gamma[6] 0.04 0.00 0.17 -0.30 -0.04 0.02 0.10 0.42 3000 1 sigma 0.40 0.00 0.05 0.32 0.36 0.40 0.43 0.51 2658 1 sigma_Z 0.19 0.00 0.18 0.02 0.07 0.14 0.24 0.65 2700 1 sd_Resid 0.39 0.00 0.01 0.37 0.38 0.39 0.39 0.42 2878 1 lp__ 22.54 0.09 4.47 13.90 19.66 22.42 25.43 31.65 2635 1 Samples were drawn using NUTS(diag_e) at Wed Jan 28 13:26:47 2015. For each parameter, n_eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence, Rhat=1).Hierarchical parameterization STAN codeprint(curdies.rstan.b)
Inference for Stan model: rstanString. 3 chains, each with iter=106000; warmup=6000; thin=100; post-warmup draws per chain=1000, total post-warmup draws=3000. mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat beta[1] 1.10 0.00 0.17 0.78 1.01 1.10 1.19 1.44 1586 1.00 beta[2] -0.80 0.01 0.29 -1.28 -0.91 -0.80 -0.67 -0.35 3000 1.00 gamma[1] 1.12 0.01 0.13 0.87 1.03 1.12 1.20 1.37 594 1.01 gamma[2] 1.17 0.00 0.14 0.91 1.08 1.17 1.26 1.46 1908 1.00 gamma[3] 1.00 0.01 0.15 0.68 0.91 1.01 1.11 1.32 216 1.01 gamma[4] 0.33 0.00 0.13 0.06 0.24 0.33 0.42 0.59 1197 1.00 gamma[5] 0.26 0.01 0.14 -0.03 0.17 0.26 0.35 0.50 445 1.01 gamma[6] 0.34 0.00 0.13 0.10 0.25 0.34 0.42 0.61 1305 1.00 sigma 0.40 0.00 0.05 0.31 0.37 0.40 0.43 0.52 2051 1.00 sigma_Z 0.18 0.00 0.21 0.02 0.06 0.12 0.23 0.65 2315 1.00 sd_Resid 0.39 0.00 0.01 0.37 0.38 0.39 0.39 0.42 3000 1.00 lp__ 22.74 0.19 4.62 13.47 19.87 22.76 25.57 31.85 599 1.00 Samples were drawn using NUTS(diag_e) at Wed Jan 28 13:32:21 2015. For each parameter, n_eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence, Rhat=1). - How about that summary figure then?
Matrix parameterization JAGS code
preds <- c('beta[1]','beta[2]') coefs <- curdies.r2jags.c$BUGSoutput$sims.matrix[,preds] curdies$SEASON <- factor(curdies$SEASON, levels=c("WINTER","SUMMER")) newdata <- data.frame(SEASON=levels(curdies$SEASON)) Xmat <- model.matrix(~SEASON, newdata) pred <- coefs %*% t(Xmat) library(plyr) newdata <- cbind(newdata, adply(pred, 2, function(x) { data.frame(Median=median(x)^4, HPDinterval(as.mcmc(x))^4, HPDinterval(as.mcmc(x), p=0.68)^4) })) newdata
SEASON X1 Median lower upper lower.1 upper.1 1 WINTER 1 0.008264 1.848e-06 0.1632 0.001182 0.04203 2 SUMMER 2 1.431345 4.249e-01 4.5684 0.832609 2.21439
library(ggplot2) p1 <- ggplot(newdata, aes(y=Median, x=SEASON)) + geom_linerange(aes(ymin=lower, ymax=upper), show_guide=FALSE)+ geom_linerange(aes(ymin=lower.1, ymax=upper.1), size=2,show_guide=FALSE)+ geom_point(size=4, shape=21, fill="white")+ scale_y_sqrt('Number of Dugesia per stone')+ theme_classic()+ theme(axis.title.y=element_text(vjust=2,size=rel(1.2)), axis.title.x=element_text(vjust=-2,size=rel(1.2)), plot.margin=unit(c(0.5,0.5,2,2), 'lines')) preds <- c('sd.Resid','sd.Site','sd.Season') curdies.sd <- adply(curdies.r2jags.c$BUGSoutput$sims.matrix[,preds],2,function(x) { data.frame(mean=mean(x), median=median(x), HPDinterval(as.mcmc(x)), HPDinterval(as.mcmc(x),p=0.68)) }) head(curdies.sd)
X1 mean median lower upper lower.1 upper.1 1 sd.Resid 0.3873 0.38542 3.629e-01 0.4146 0.369787 0.3941 2 sd.Site 0.1047 0.09569 4.515e-05 0.2360 0.002005 0.1334 3 sd.Season 0.5636 0.55946 2.345e-01 0.8925 0.431956 0.6992
rownames(curdies.sd) <- c("Residuals", "Site", "Season") curdies.sd$name <- factor(c("Residuals", "Site", "Season"), levels=c("Residuals", "Site", "Season")) curdies.sd$Perc <- curdies.sd$median/sum(curdies.sd$median) p2<-ggplot(curdies.sd,aes(y=name, x=median))+ geom_vline(xintercept=0,linetype="dashed")+ geom_hline(xintercept=0)+ scale_x_continuous("Finite population \nvariance components (sd)")+ geom_errorbarh(aes(xmin=lower.1, xmax=upper.1), height=0, size=1)+ geom_errorbarh(aes(xmin=lower, xmax=upper), height=0, size=1.5)+ geom_point(size=3, shape=21, fill='white')+ geom_text(aes(label=sprintf("(%4.1f%%)",Perc),vjust=-1))+ theme_classic()+ theme(axis.title.y=element_blank(), axis.text.y=element_text(size=rel(1.2),hjust=1)) library(gridExtra) grid.arrange(p1,p2,ncol=2)
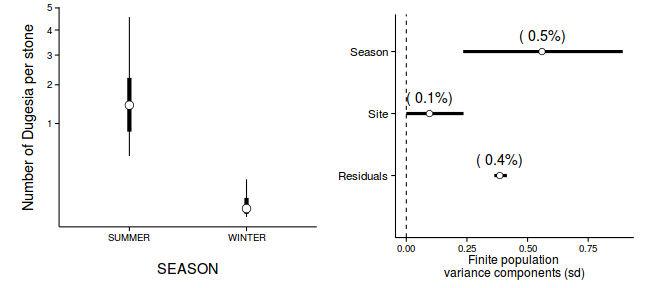
Exploratory data analysis has indicated that the response variable could be normalized via a forth-root transformation.
