Workshop 9.4b - Mixed effects (Multilevel models): Split-plot and complex repeated measures designs (Bayesian)
31 Jan 2015
Split-plot ANOVA (Mixed effects) references
- McCarthy (2007) - Chpt ?
- Kery (2010) - Chpt ?
- Gelman & Hill (2007) - Chpt ?
- Logan (2010) - Chpt 13
- Quinn & Keough (2002) - Chpt 10
Split-plot
Split-plotIn an attempt to understand the effects on marine animals of short-term exposure to toxic substances, such as might occur following a spill, or a major increase in storm water flows, a it was decided to examine the toxicant in question, Copper, as part of a field experiment in Honk Kong. The experiment consisted of small sources of Cu (small, hemispherical plaster blocks, impregnated with copper), which released the metal into sea water over 4 or 5 days. The organism whose response to Cu was being measured was a small, polychaete worm, Hydroides, that attaches to hard surfaces in the sea, and is one of the first species to colonize any surface that is submerged. The biological questions focused on whether the timing of exposure to Cu affects the overall abundance of these worms. The time period of interest was the first or second week after a surface being available.
The experimental setup consisted of sheets of black perspex (settlement plates), which provided good surfaces for these worms. Each plate had a plaster block bolted to its centre, and the dissolving block would create a gradient of [Cu] across the plate. Over the two weeks of the experiment, a given plate would have pl ain plaster blocks (Control) or a block containing copper in the first week, followed by a plain block, or a plain block in the first week, followed by a dose of copper in the second week. After two weeks in the water, plates were removed and counted back in the laboratory. Without a clear idea of how sensitive these worms are to copper, an effect of the treatments might show up as an overall difference in the density of worms across a plate, or it could show up as a gradient in abundance across the plate, with a different gradient in different treatments. Therefore, on each plate, the density of worms (#/cm2) was recorded at each of four distances from the center of the plate.
Download Copper data set| Format of copper.csv data file | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
 |
copper <- read.table('../downloads/data/copper.csv', header=T, sep=',', strip.white=T) head(copper)
COPPER PLATE DIST WORMS 1 control 200 4 11.50 2 control 200 3 13.00 3 control 200 2 13.50 4 control 200 1 12.00 5 control 39 4 17.75 6 control 39 3 13.75
library(R2jags) library(rstan) library(coda)
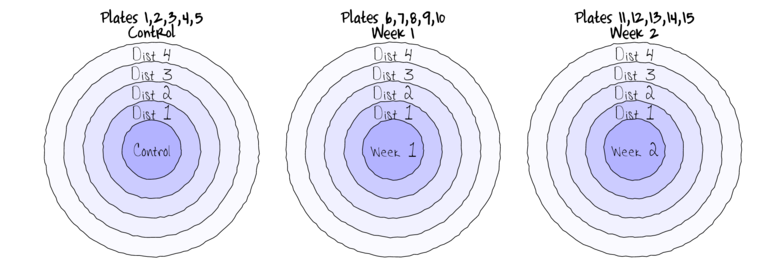
The Plates are the "random" groups. Within each Plate, all levels of the Distance factor occur (this is a within group factor). Each Plate can only be of one of the three levels of the Copper treatment. This is therefore a within group (nested) factor. Traditionally, this mixture of nested and randomized block design would be called a partly nested or split-plot design.
In Bayesian (multilevel modeling) terms, this is a multi-level model with one hierarchical level the Plates means and another representing the Copper treatment means (based on the Plate means).
Exploratory data analysis has indicated that the response variable could be normalized via a forth-root transformation.
- Fit a model for Randomized Complete Block
- Effects parameterization - random intercepts model (JAGS)
number of lesionsi = βSite j(i) + εi where ε ∼ N(0,σ2)treating Distance as a factor
View full codemodelString= ## write the model to a text file (I suggest you alter the path to somewhere more relevant ## to your system!) writeLines(modelString,con="../downloads/BUGSscripts/ws9.9bQ2.1a.txt")
#sort the data set so that the copper treatments are in a more logical order copper.sort <- copper[order(copper$COPPER),] copper.sort$COPPER <- factor(copper.sort$COPPER, levels = c("control", "Week 1", "Week 2")) copper.sort$PLATE <- factor(copper.sort$PLATE, levels=unique(copper.sort$PLATE)) cu <- with(copper.sort,COPPER[match(unique(PLATE),PLATE)]) cu <- factor(cu, levels=c("control", "Week 1", "Week 2")) copper.list <- with(copper.sort, list(worms=WORMS, dist=as.numeric(DIST), plate=as.numeric(PLATE), copper=factor(as.numeric(cu)), cu=as.numeric(COPPER), nDist=length(levels(as.factor(DIST))), nPlate=length(levels(as.factor(PLATE))), nCopper=length(levels(cu)), n=nrow(copper.sort) ) ) params <- c("g0","gamma.copper", "beta.plate", "beta.dist", "beta.CopperDist", "Copper.means","Dist.means","CopperDist.means", "sd.Dist","sd.Copper","sd.Plate","sd.Resid","sd.CopperDist", "sigma.res","sigma.plate","sigma.copper","sigma.dist","sigma.copperdist" ) adaptSteps = 1000 burnInSteps = 2000 nChains = 3 numSavedSteps = 5000 thinSteps = 10 nIter = ceiling((numSavedSteps * thinSteps)/nChains) library(R2jags) ## copper.r2jags.a <- jags(data=copper.list, inits=NULL, # since there are three chains parameters.to.save=params, model.file="../downloads/BUGSscripts/ws9.9bQ2.1a.txt", n.chains=3, n.iter=nIter, n.burnin=burnInSteps, n.thin=thinSteps )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 458 Initializing model
print(copper.r2jags.a)
Inference for Bugs model at "../downloads/BUGSscripts/ws9.9bQ2.1a.txt", fit using jags, 3 chains, each with 16667 iterations (first 2000 discarded), n.thin = 10 n.sims = 4401 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff Copper.means[1] 10.853 0.720 9.410 10.381 10.859 11.321 12.235 1.001 3700 Copper.means[2] 6.966 0.684 5.671 6.517 6.967 7.412 8.318 1.001 3800 Copper.means[3] 0.493 0.690 -0.874 0.037 0.499 0.947 1.839 1.002 1500 CopperDist.means[1,1] 10.853 0.720 9.410 10.381 10.859 11.321 12.235 1.001 3700 CopperDist.means[2,1] 6.966 0.684 5.671 6.517 6.967 7.412 8.318 1.001 3800 CopperDist.means[3,1] 0.493 0.690 -0.874 0.037 0.499 0.947 1.839 1.002 1500 CopperDist.means[1,2] 12.008 0.676 10.668 11.563 12.018 12.459 13.309 1.001 3500 CopperDist.means[2,2] 8.405 0.674 7.055 7.963 8.406 8.854 9.687 1.001 4400 CopperDist.means[3,2] 1.579 0.679 0.255 1.121 1.584 2.026 2.908 1.001 4400 CopperDist.means[1,3] 12.428 0.642 11.152 12.018 12.414 12.838 13.708 1.001 2700 CopperDist.means[2,3] 8.591 0.683 7.245 8.135 8.586 9.052 9.951 1.001 4400 CopperDist.means[3,3] 3.932 0.681 2.586 3.485 3.920 4.380 5.298 1.001 4400 CopperDist.means[1,4] 13.523 0.708 12.190 13.032 13.531 13.989 14.896 1.001 2500 CopperDist.means[2,4] 10.088 0.676 8.750 9.641 10.078 10.546 11.434 1.001 4400 CopperDist.means[3,4] 7.543 0.720 6.117 7.056 7.557 8.034 8.912 1.001 4400 Dist.means[1] 6.107 0.366 5.367 5.869 6.115 6.340 6.827 1.002 4400 Dist.means[2] 7.262 0.798 5.725 6.724 7.267 7.800 8.811 1.001 4400 Dist.means[3] 7.682 0.779 6.113 7.172 7.691 8.187 9.228 1.001 4400 Dist.means[4] 8.777 0.826 7.159 8.223 8.754 9.324 10.417 1.001 3500 beta.CopperDist[1,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.CopperDist[2,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.CopperDist[3,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.CopperDist[1,2] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.CopperDist[2,2] 0.284 1.240 -2.158 -0.561 0.274 1.106 2.751 1.001 4400 beta.CopperDist[3,2] -0.068 1.207 -2.480 -0.874 -0.078 0.760 2.271 1.001 3700 beta.CopperDist[1,3] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.CopperDist[2,3] 0.050 1.214 -2.305 -0.756 0.051 0.841 2.413 1.001 3900 beta.CopperDist[3,3] 1.865 1.198 -0.556 1.074 1.873 2.670 4.279 1.001 3300 beta.CopperDist[1,4] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.CopperDist[2,4] 0.452 1.206 -1.862 -0.367 0.463 1.272 2.824 1.001 4100 beta.CopperDist[3,4] 4.381 1.332 1.695 3.507 4.404 5.274 6.975 1.002 1700 beta.dist[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.dist[2] 1.155 0.875 -0.570 0.573 1.158 1.737 2.857 1.001 4400 beta.dist[3] 1.575 0.874 -0.170 1.001 1.585 2.135 3.320 1.001 4400 beta.dist[4] 2.670 0.911 0.926 2.069 2.660 3.274 4.489 1.001 4400 beta.plate[1] 10.968 0.761 9.466 10.450 10.967 11.482 12.429 1.001 4400 beta.plate[2] 11.627 0.875 9.934 11.039 11.618 12.213 13.359 1.001 3400 beta.plate[3] 10.449 0.786 8.825 9.918 10.476 10.998 11.951 1.001 4400 beta.plate[4] 10.505 0.784 8.885 10.000 10.528 11.031 12.011 1.001 4400 beta.plate[5] 10.728 0.759 9.153 10.242 10.743 11.233 12.223 1.002 4400 beta.plate[6] 6.861 0.720 5.438 6.385 6.857 7.329 8.312 1.001 4400 beta.plate[7] 6.505 0.763 4.948 6.012 6.509 7.014 7.976 1.001 4400 beta.plate[8] 7.245 0.741 5.845 6.743 7.233 7.753 8.734 1.001 2900 beta.plate[9] 7.107 0.724 5.709 6.613 7.100 7.582 8.560 1.001 4400 beta.plate[10] 7.129 0.736 5.700 6.640 7.123 7.618 8.582 1.001 4400 beta.plate[11] 0.420 0.736 -1.026 -0.071 0.421 0.910 1.876 1.001 2500 beta.plate[12] 0.343 0.737 -1.091 -0.146 0.340 0.848 1.790 1.003 980 beta.plate[13] 0.387 0.737 -1.022 -0.108 0.393 0.879 1.808 1.002 1300 beta.plate[14] 0.707 0.738 -0.735 0.215 0.709 1.188 2.144 1.001 3500 beta.plate[15] 0.621 0.736 -0.835 0.135 0.623 1.109 2.066 1.002 1700 g0 10.853 0.720 9.410 10.381 10.859 11.321 12.235 1.001 3700 gamma.copper[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 gamma.copper[2] -3.886 0.986 -5.802 -4.551 -3.875 -3.221 -1.907 1.002 1800 gamma.copper[3] -10.360 0.995 -12.307 -11.007 -10.357 -9.709 -8.362 1.001 3900 sd.Copper 5.256 0.495 4.244 4.936 5.251 5.578 6.246 1.001 2800 sd.CopperDist 1.484 0.351 0.817 1.248 1.477 1.714 2.202 1.001 3500 sd.Dist 1.208 0.360 0.511 0.962 1.205 1.448 1.937 1.001 4400 sd.Plate 0.528 0.271 0.045 0.326 0.528 0.724 1.048 1.004 1400 sd.Resid 4.304 0.000 4.304 4.304 4.304 4.304 4.304 1.000 1 sigma.copper 29.628 25.472 2.924 8.959 20.406 44.181 91.539 1.001 2800 sigma.copperdist 2.660 1.611 0.912 1.690 2.263 3.160 6.733 1.001 3200 sigma.dist 5.912 12.445 0.133 0.880 1.769 4.373 47.253 1.015 300 sigma.plate 0.587 0.334 0.050 0.343 0.560 0.795 1.318 1.003 1700 sigma.res 1.408 0.166 1.121 1.289 1.398 1.511 1.761 1.002 2300 deviance 209.192 8.606 193.679 203.023 208.938 214.934 226.967 1.002 1400 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 37.0 and DIC = 246.2 DIC is an estimate of expected predictive error (lower deviance is better). - Matrix parameterization - random intercepts model (JAGS)
number of lesionsi = βSite j(i) + εi where ε ∼ N(0,σ2)treating Distance as a factor
View full codeWe can perform generate the finite-population standard deviation posteriors within RmodelString=" model { #Likelihood for (i in 1:n) { y[i]~dnorm(mu[i],tau.res) mu[i] <- inprod(beta[],X[i,]) + inprod(gamma[],Z[i,]) y.err[i] <- y[i] - mu[1] } #Priors and derivatives for (i in 1:nZ) { gamma[i] ~ dnorm(0,tau.plate) } for (i in 1:nX) { beta[i] ~ dnorm(0,1.0E-06) } tau.res <- pow(sigma.res,-2) sigma.res <- z/sqrt(chSq) z ~ dnorm(0, .0016)I(0,) chSq ~ dgamma(0.5, 0.5) tau.plate <- pow(sigma.plate,-2) sigma.plate <- z.plate/sqrt(chSq.plate) z.plate ~ dnorm(0, .0016)I(0,) chSq.plate ~ dgamma(0.5, 0.5) } " ## write the model to a text file (I suggest you alter the path to somewhere more relevant ## to your system!) writeLines(modelString,con="../downloads/BUGSscripts/ws9.4bQ2.1b.txt") #sort the data set so that the copper treatments are in a more logical order library(dplyr) copper$DIST <- factor(copper$DIST) copper$PLATE <- factor(copper$PLATE) copper.sort <- arrange(copper,COPPER,PLATE,DIST) Xmat <- model.matrix(~COPPER*DIST, data=copper.sort) Zmat <- model.matrix(~-1+PLATE, data=copper.sort) copper.list <- list(y=copper.sort$WORMS, X=Xmat, nX=ncol(Xmat), Z=Zmat, nZ=ncol(Zmat), n=nrow(copper.sort) ) params <- c("beta","gamma", "sigma.res","sigma.plate") burnInSteps = 3000 nChains = 3 numSavedSteps = 3000 thinSteps = 100 nIter = ceiling((numSavedSteps * thinSteps)/nChains) library(R2jags) library(coda) ## copper.r2jags.b <- jags(data=copper.list, inits=NULL, # since there are three chains parameters.to.save=params, model.file="../downloads/BUGSscripts/ws9.4bQ2.1b.txt", n.chains=3, n.iter=nIter, n.burnin=burnInSteps, n.thin=thinSteps )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 1971 Initializing model
print(copper.r2jags.b)
Inference for Bugs model at "../downloads/BUGSscripts/ws9.4bQ2.1b.txt", fit using jags, 3 chains, each with 1e+05 iterations (first 3000 discarded), n.thin = 100 n.sims = 2910 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff beta[1] 10.849 0.684 9.531 10.377 10.857 11.304 12.152 1.002 1200 beta[2] -3.617 0.977 -5.522 -4.272 -3.608 -2.979 -1.701 1.003 740 beta[3] -10.573 0.976 -12.521 -11.215 -10.588 -9.909 -8.713 1.002 1900 beta[4] 1.161 0.877 -0.575 0.588 1.157 1.759 2.852 1.001 2900 beta[5] 1.552 0.885 -0.147 0.950 1.550 2.160 3.270 1.003 690 beta[6] 2.712 0.894 0.923 2.118 2.715 3.286 4.471 1.001 2900 beta[7] -0.041 1.274 -2.579 -0.876 -0.004 0.828 2.370 1.001 2900 beta[8] 0.007 1.241 -2.452 -0.810 -0.007 0.831 2.453 1.001 2900 beta[9] -0.264 1.237 -2.641 -1.084 -0.241 0.529 2.223 1.002 1300 beta[10] 2.175 1.244 -0.309 1.332 2.181 2.953 4.666 1.001 2900 beta[11] 0.066 1.279 -2.410 -0.760 0.080 0.889 2.605 1.001 2900 beta[12] 4.870 1.260 2.376 4.055 4.890 5.696 7.346 1.011 2900 gamma[1] 0.156 0.490 -0.774 -0.122 0.095 0.419 1.276 1.001 2900 gamma[2] -0.140 0.492 -1.214 -0.407 -0.088 0.141 0.805 1.001 2900 gamma[3] 0.285 0.511 -0.606 -0.029 0.201 0.581 1.435 1.001 2900 gamma[4] -0.423 0.547 -1.665 -0.732 -0.335 -0.030 0.469 1.001 2900 gamma[5] -0.360 0.531 -1.518 -0.655 -0.275 -0.006 0.525 1.002 1300 gamma[6] 0.765 0.664 -0.170 0.217 0.677 1.193 2.238 1.001 2900 gamma[7] -0.153 0.493 -1.219 -0.430 -0.091 0.117 0.781 1.003 850 gamma[8] -0.491 0.558 -1.786 -0.829 -0.397 -0.067 0.370 1.001 2900 gamma[9] 0.138 0.497 -0.810 -0.145 0.082 0.412 1.255 1.001 2900 gamma[10] -0.106 0.486 -1.120 -0.386 -0.061 0.160 0.877 1.002 1500 gamma[11] -0.148 0.495 -1.204 -0.415 -0.096 0.128 0.816 1.002 1100 gamma[12] 0.213 0.503 -0.710 -0.076 0.152 0.488 1.308 1.003 760 gamma[13] 0.118 0.488 -0.886 -0.145 0.079 0.388 1.178 1.002 2200 gamma[14] 0.121 0.492 -0.871 -0.145 0.081 0.385 1.185 1.001 2900 gamma[15] -0.092 0.502 -1.197 -0.359 -0.055 0.183 0.902 1.002 1800 sigma.plate 0.588 0.328 0.044 0.352 0.572 0.788 1.287 1.003 1600 sigma.res 1.397 0.164 1.102 1.284 1.386 1.502 1.745 1.001 2900 deviance 208.815 8.504 193.162 202.760 208.517 214.590 226.162 1.001 2300 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 36.2 and DIC = 245.0 DIC is an estimate of expected predictive error (lower deviance is better).copper.mcmc.b <- as.mcmc(copper.r2jags.b$BUGSoutput$sims.matrix)
View full codecopper.sort <- arrange(copper,COPPER,PLATE,DIST) nms <- colnames(copper.mcmc.b) pred <- grep('beta|gamma',nms) coefs <- copper.mcmc.b[,pred] newdata <- copper.sort #unique(subset(copper, select=c('COPPER','DIST','PLATE'))) Amat <- cbind(model.matrix(~COPPER*DIST, newdata),model.matrix(~-1+PLATE, newdata)) pred <- coefs %*% t(Amat) y.err <- t(copper.sort$WORMS - t(pred)) a<-apply(y.err, 1,sd) resid.sd <- data.frame(Source='Resid',Median=median(a), HPDinterval(as.mcmc(a)), HPDinterval(as.mcmc(a), p=0.68)) ## Fixed effects nms <- colnames(copper.mcmc.b) copper.mcmc.b.beta <- copper.mcmc.b[,grep('beta',nms)] assign <- attr(Xmat, 'assign') nms <- attr(terms(model.frame(~COPPER*DIST, copper.sort)),'term.labels') fixed.sd <- NULL for (i in 1:max(assign)) { w <- which(i==assign) fixed.sd<-cbind(fixed.sd, apply(copper.mcmc.b.beta[,w],1,sd)) } library(plyr) fixed.sd <-adply(fixed.sd,2,function(x) { data.frame(Median=median(x), HPDinterval(as.mcmc(x)), HPDinterval(as.mcmc(x), p=0.68)) }) fixed.sd<- cbind(Source=nms, fixed.sd) # Variances nms <- colnames(copper.mcmc.b) copper.mcmc.b.gamma <- copper.mcmc.b[,grep('gamma',nms)] assign <- attr(Zmat, 'assign') nms <- attr(terms(model.frame(~-1+PLATE, copper.sort)),'term.labels') random.sd <- NULL for (i in 1:max(assign)) { w <- which(i==assign) random.sd<-cbind(random.sd, apply(copper.mcmc.b.gamma[,w],1,sd)) } library(plyr) random.sd <-adply(random.sd,2,function(x) { data.frame(Median=median(x), HPDinterval(as.mcmc(x)), HPDinterval(as.mcmc(x), p=0.68)) }) random.sd<- cbind(Source=nms, random.sd) (copper.sd <- rbind(subset(fixed.sd, select=-X1), subset(random.sd, select=-X1), resid.sd))
Source Median lower upper lower.1 upper.1 1 COPPER 4.9703 3.6177274 6.4345 4.2476 5.6256 2 DIST 0.9303 0.1402592 1.6415 0.5160 1.3082 3 COPPER:DIST 2.2198 1.3338084 3.1178 1.7949 2.6786 4 PLATE 0.5333 0.0002546 0.9595 0.2442 0.7985 var1 Resid 1.3648 1.1950027 1.5576 1.2624 1.4503
- Matrix parameterization - random intercepts model (STAN)
View full coderstanString=" data{ int n; int nZ; int nX; vector [n] y; matrix [n,nX] X; matrix [n,nZ] Z; vector [nX] a0; matrix [nX,nX] A0; } parameters{ vector [nX] beta; real<lower=0> sigma; vector [nZ] gamma; real<lower=0> sigma_Z; } transformed parameters { vector [n] mu; mu <- X*beta + Z*gamma; } model{ // Priors beta ~ multi_normal(a0,A0); gamma ~ normal( 0 , sigma_Z ); sigma_Z ~ cauchy(0,25); sigma ~ cauchy(0,25); y ~ normal( mu , sigma ); } generated quantities { vector [n] y_err; real<lower=0> sd_Resid; y_err <- y - mu; sd_Resid <- sd(y_err); } " #sort the data set so that the copper treatments are in a more logical order library(dplyr) copper$DIST <- factor(copper$DIST) copper$PLATE <- factor(copper$PLATE) copper.sort <- arrange(copper,COPPER,PLATE,DIST) Xmat <- model.matrix(~COPPER*DIST, data=copper.sort) Zmat <- model.matrix(~-1+PLATE, data=copper.sort) copper.list <- list(y=copper.sort$WORMS, X=Xmat, nX=ncol(Xmat), Z=Zmat, nZ=ncol(Zmat), n=nrow(copper.sort), a0=rep(0,ncol(Xmat)), A0=diag(100000,ncol(Xmat)) ) params <- c("beta","gamma", "sigma","sigma_Z") burnInSteps = 3000 nChains = 3 numSavedSteps = 3000 thinSteps = 100 nIter = ceiling((numSavedSteps * thinSteps)/nChains) library(rstan) library(coda) ## copper.rstan.a <- stan(data=copper.list, model_code=rstanString, pars=c("beta","gamma","sigma","sigma_Z"), chains=nChains, iter=nIter, warmup=burnInSteps, thin=thinSteps, save_dso=TRUE )
TRANSLATING MODEL 'rstanString' FROM Stan CODE TO C++ CODE NOW. COMPILING THE C++ CODE FOR MODEL 'rstanString' NOW. SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 1). Iteration: 1 / 100000 [ 0%] (Warmup) Iteration: 3001 / 100000 [ 3%] (Sampling) Iteration: 13000 / 100000 [ 13%] (Sampling) Iteration: 23000 / 100000 [ 23%] (Sampling) Iteration: 33000 / 100000 [ 33%] (Sampling) Iteration: 43000 / 100000 [ 43%] (Sampling) Iteration: 53000 / 100000 [ 53%] (Sampling) Iteration: 63000 / 100000 [ 63%] (Sampling) Iteration: 73000 / 100000 [ 73%] (Sampling) Iteration: 83000 / 100000 [ 83%] (Sampling) Iteration: 93000 / 100000 [ 93%] (Sampling) Iteration: 100000 / 100000 [100%] (Sampling) # Elapsed Time: 1.58 seconds (Warm-up) # 64.79 seconds (Sampling) # 66.37 seconds (Total) SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 2). Iteration: 1 / 100000 [ 0%] (Warmup) Iteration: 3001 / 100000 [ 3%] (Sampling) Iteration: 13000 / 100000 [ 13%] (Sampling) Iteration: 23000 / 100000 [ 23%] (Sampling) Iteration: 33000 / 100000 [ 33%] (Sampling) Iteration: 43000 / 100000 [ 43%] (Sampling) Iteration: 53000 / 100000 [ 53%] (Sampling) Iteration: 63000 / 100000 [ 63%] (Sampling) Iteration: 73000 / 100000 [ 73%] (Sampling) Iteration: 83000 / 100000 [ 83%] (Sampling) Iteration: 93000 / 100000 [ 93%] (Sampling) Iteration: 100000 / 100000 [100%] (Sampling) # Elapsed Time: 1.52 seconds (Warm-up) # 73.33 seconds (Sampling) # 74.85 seconds (Total) SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 3). Iteration: 1 / 100000 [ 0%] (Warmup) Iteration: 3001 / 100000 [ 3%] (Sampling) Iteration: 13000 / 100000 [ 13%] (Sampling) Iteration: 23000 / 100000 [ 23%] (Sampling) Iteration: 33000 / 100000 [ 33%] (Sampling) Iteration: 43000 / 100000 [ 43%] (Sampling) Iteration: 53000 / 100000 [ 53%] (Sampling) Iteration: 63000 / 100000 [ 63%] (Sampling) Iteration: 73000 / 100000 [ 73%] (Sampling) Iteration: 83000 / 100000 [ 83%] (Sampling) Iteration: 93000 / 100000 [ 93%] (Sampling) Iteration: 100000 / 100000 [100%] (Sampling) # Elapsed Time: 2.35 seconds (Warm-up) # 69.92 seconds (Sampling) # 72.27 seconds (Total)
print(copper.rstan.a)
Inference for Stan model: rstanString. 3 chains, each with iter=1e+05; warmup=3000; thin=100; post-warmup draws per chain=970, total post-warmup draws=2910. mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat beta[1] 10.86 0.01 0.69 9.49 10.40 10.86 11.34 12.21 2910 1 beta[2] -3.59 0.02 0.98 -5.49 -4.28 -3.59 -2.93 -1.70 2670 1 beta[3] -10.58 0.02 0.98 -12.51 -11.23 -10.58 -9.93 -8.63 2910 1 beta[4] 1.15 0.02 0.91 -0.66 0.54 1.16 1.75 2.95 2672 1 beta[5] 1.56 0.02 0.89 -0.22 0.96 1.56 2.12 3.35 2909 1 beta[6] 2.69 0.02 0.90 0.94 2.10 2.69 3.28 4.50 2807 1 beta[7] -0.06 0.02 1.29 -2.58 -0.92 -0.08 0.80 2.47 2670 1 beta[8] 0.03 0.02 1.27 -2.52 -0.80 0.02 0.86 2.50 2910 1 beta[9] -0.30 0.02 1.27 -2.74 -1.16 -0.31 0.54 2.24 2910 1 beta[10] 2.19 0.02 1.25 -0.26 1.37 2.20 3.03 4.60 2910 1 beta[11] 0.06 0.02 1.27 -2.49 -0.78 0.06 0.91 2.51 2892 1 beta[12] 4.88 0.02 1.25 2.32 4.06 4.92 5.70 7.26 2910 1 gamma[1] 0.15 0.01 0.49 -0.80 -0.12 0.11 0.39 1.21 2910 1 gamma[2] -0.13 0.01 0.50 -1.23 -0.40 -0.09 0.14 0.85 2880 1 gamma[3] 0.27 0.01 0.51 -0.63 -0.05 0.19 0.54 1.43 2910 1 gamma[4] -0.42 0.01 0.55 -1.66 -0.73 -0.32 -0.03 0.46 2910 1 gamma[5] -0.36 0.01 0.55 -1.56 -0.68 -0.28 0.00 0.60 2910 1 gamma[6] 0.77 0.01 0.67 -0.18 0.23 0.69 1.19 2.26 2861 1 gamma[7] -0.16 0.01 0.50 -1.29 -0.44 -0.10 0.13 0.79 2910 1 gamma[8] -0.49 0.01 0.56 -1.78 -0.83 -0.40 -0.08 0.37 2910 1 gamma[9] 0.14 0.01 0.50 -0.82 -0.14 0.10 0.41 1.20 2910 1 gamma[10] -0.12 0.01 0.50 -1.22 -0.38 -0.07 0.16 0.83 2910 1 gamma[11] -0.13 0.01 0.50 -1.25 -0.39 -0.09 0.14 0.81 2910 1 gamma[12] 0.20 0.01 0.50 -0.71 -0.10 0.14 0.49 1.33 2910 1 gamma[13] 0.12 0.01 0.50 -0.86 -0.15 0.08 0.39 1.23 2648 1 gamma[14] 0.12 0.01 0.49 -0.85 -0.14 0.08 0.37 1.21 2895 1 gamma[15] -0.08 0.01 0.51 -1.18 -0.34 -0.04 0.20 0.93 2884 1 sigma 1.40 0.00 0.16 1.13 1.29 1.39 1.50 1.75 2811 1 sigma_Z 0.59 0.01 0.33 0.07 0.35 0.57 0.79 1.32 2899 1 lp__ -45.79 0.16 8.69 -59.87 -51.27 -46.89 -41.98 -23.18 2910 1 Samples were drawn using NUTS(diag_e) at Fri Jan 30 14:27:46 2015. For each parameter, n_eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence, Rhat=1).copper.mcmc.c <- rstan:::as.mcmc.list.stanfit(copper.rstan.a) copper.mcmc.df.c <- as.data.frame(extract(copper.rstan.a))
- Effects parameterization - random intercepts model (JAGS)
- Before fully exploring the parameters, it is prudent to examine the convergence and mixing diagnostics.
Chose either any of the parameterizations (they should yield much the same) - although it is sometimes useful to explore the
different performances of effects vs matrix and JAGS vs STAN.
- Effects parameterization - random intercepts model (JAGS)
Full effects parameterization JAGS code
library(coda) plot(as.mcmc(copper.r2jags.a))

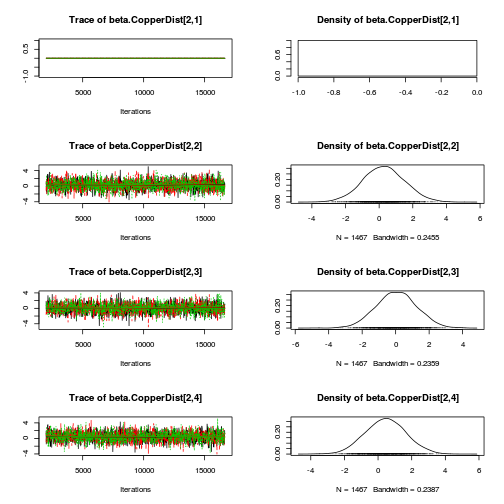
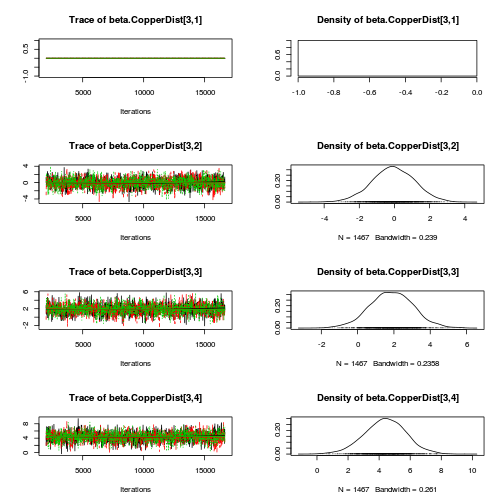
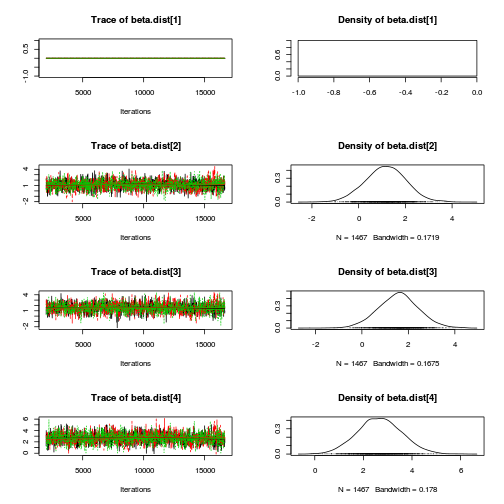
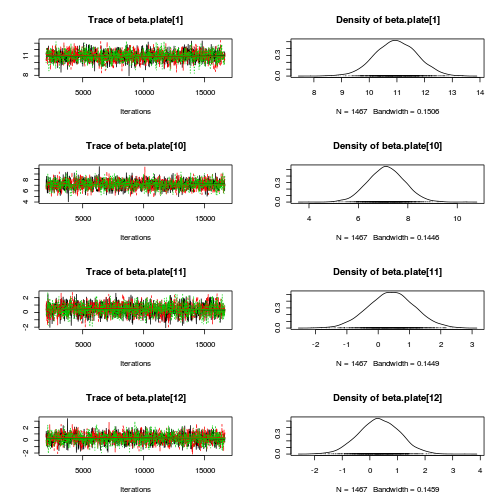
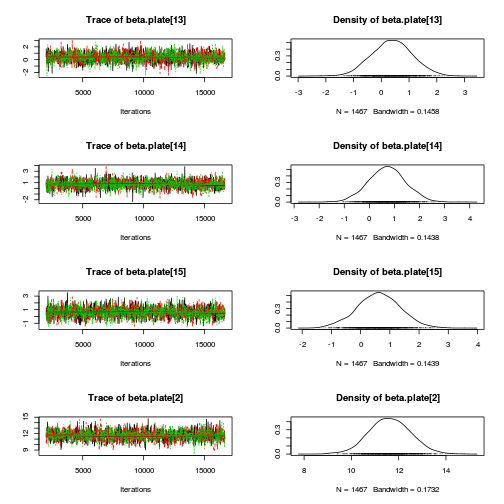
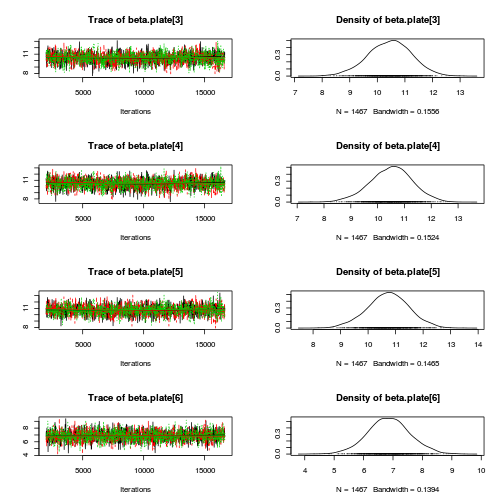
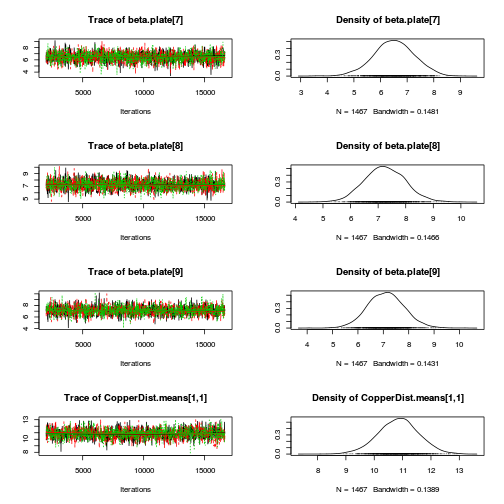
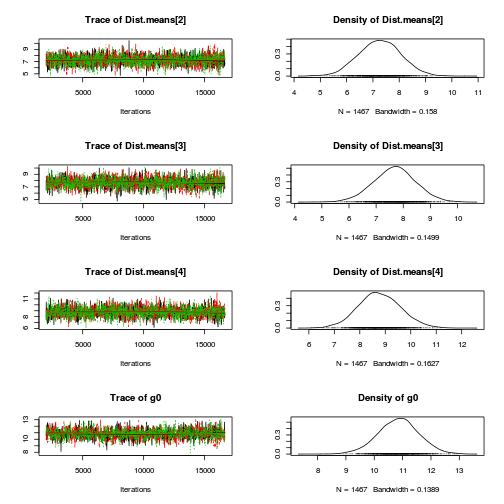
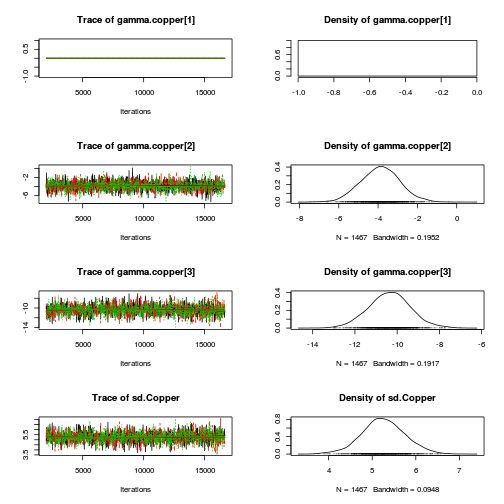
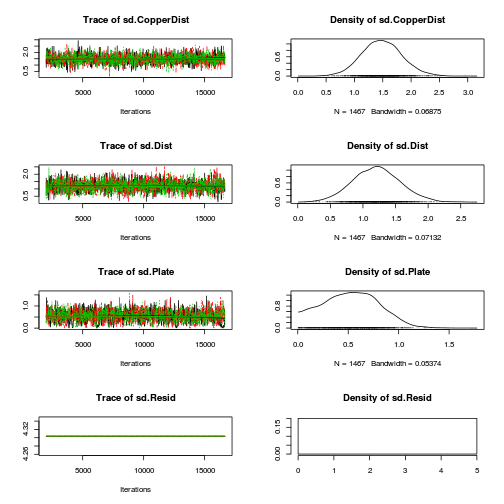
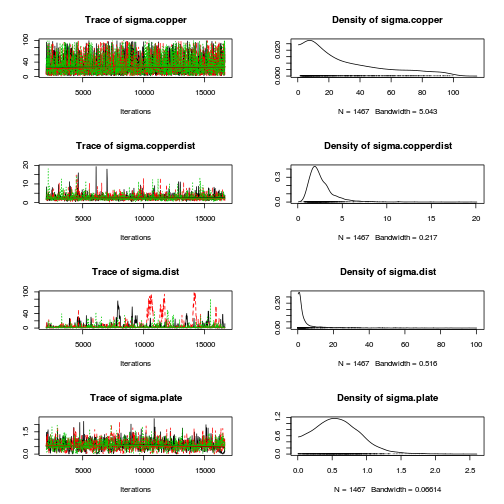
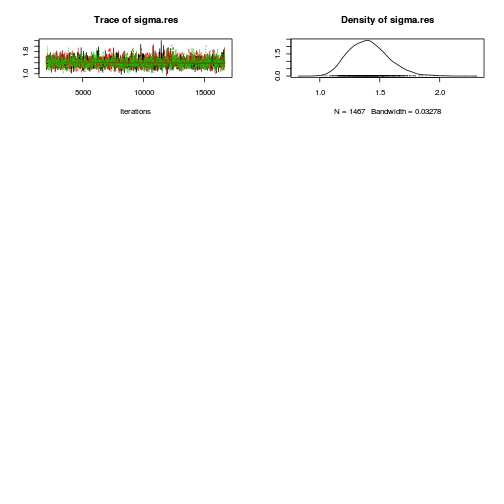
autocorr.diag(as.mcmc(copper.r2jags.a))
beta.CopperDist[1,1] beta.CopperDist[1,2] beta.CopperDist[1,3] beta.CopperDist[1,4] Lag 0 NaN NaN NaN NaN Lag 10 NaN NaN NaN NaN Lag 50 NaN NaN NaN NaN Lag 100 NaN NaN NaN NaN Lag 500 NaN NaN NaN NaN beta.CopperDist[2,1] beta.CopperDist[2,2] beta.CopperDist[2,3] beta.CopperDist[2,4] Lag 0 NaN 1.000000 1.000000 1.000000 Lag 10 NaN 0.253736 0.227276 0.203586 Lag 50 NaN 0.035243 0.040309 0.021789 Lag 100 NaN 0.004880 0.006712 0.006038 Lag 500 NaN -0.005862 0.009423 -0.019633 beta.CopperDist[3,1] beta.CopperDist[3,2] beta.CopperDist[3,3] beta.CopperDist[3,4] Lag 0 NaN 1.00000 1.000000 1.00000 Lag 10 NaN 0.23379 0.229879 0.23285 Lag 50 NaN 0.05883 0.068947 0.05358 Lag 100 NaN 0.01468 0.004564 0.01374 Lag 500 NaN -0.02168 0.009124 -0.00258 beta.dist[1] beta.dist[2] beta.dist[3] beta.dist[4] beta.plate[1] beta.plate[10] Lag 0 NaN 1.00000 1.000000 1.000000 1.0000000 1.000000 Lag 10 NaN 0.28216 0.270097 0.246958 0.2877373 0.245976 Lag 50 NaN 0.04596 0.062366 0.042753 0.0512777 0.013843 Lag 100 NaN 0.02506 -0.005532 0.043504 0.0160756 0.002873 Lag 500 NaN -0.01090 0.014212 0.004496 -0.0008968 -0.002487 beta.plate[11] beta.plate[12] beta.plate[13] beta.plate[14] beta.plate[15] beta.plate[2] Lag 0 1.000000 1.00000 1.0000000 1.00000 1.00000 1.000000 Lag 10 0.265392 0.26622 0.2627529 0.23865 0.26547 0.371823 Lag 50 0.058535 0.03400 0.0467393 0.04584 0.04345 0.047311 Lag 100 0.011697 0.01575 0.0076997 0.01527 0.03520 0.000455 Lag 500 -0.002267 0.01265 -0.0001311 0.03240 0.00446 -0.015496 beta.plate[3] beta.plate[4] beta.plate[5] beta.plate[6] beta.plate[7] beta.plate[8] Lag 0 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 Lag 10 0.346744 0.316141 0.298380 0.261183 0.274941 0.273801 Lag 50 0.073440 0.082624 0.073615 0.025826 0.033300 0.036737 Lag 100 0.034460 0.036281 0.041138 0.006353 0.027049 -0.026790 Lag 500 0.003324 0.003008 -0.008167 -0.017742 -0.005289 0.009593 beta.plate[9] CopperDist.means[1,1] CopperDist.means[1,2] CopperDist.means[1,3] Lag 0 1.000000 1.000000 1.000000 1.000000 Lag 10 0.250768 0.365289 0.023337 0.028161 Lag 50 0.027479 0.074439 0.021650 0.026946 Lag 100 -0.003263 0.028816 0.011464 -0.002759 Lag 500 -0.012326 -0.005242 0.005228 -0.003432 CopperDist.means[1,4] CopperDist.means[2,1] CopperDist.means[2,2] CopperDist.means[2,3] Lag 0 1.000000 1.000000 1.000000 1.000000 Lag 10 0.038681 0.306616 0.011700 0.004240 Lag 50 0.020680 0.031770 0.014968 -0.013660 Lag 100 0.024175 0.011743 -0.004706 -0.001578 Lag 500 -0.009238 0.004162 0.010357 -0.014052 CopperDist.means[2,4] CopperDist.means[3,1] CopperDist.means[3,2] CopperDist.means[3,3] Lag 0 1.00000 1.000000 1.00000 1.000000 Lag 10 0.03281 0.316645 0.02477 0.004322 Lag 50 0.01864 0.056475 0.01592 0.003629 Lag 100 0.01215 0.022872 -0.01715 0.017472 Lag 500 0.01278 0.006836 -0.01798 0.007568 CopperDist.means[3,4] Copper.means[1] Copper.means[2] Copper.means[3] deviance Lag 0 1.000000 1.000000 1.000000 1.000000 1.00000 Lag 10 -0.016148 0.365289 0.306616 0.316645 0.19209 Lag 50 0.010683 0.074439 0.031770 0.056475 0.05804 Lag 100 -0.017415 0.028816 0.011743 0.022872 -0.02862 Lag 500 0.004738 -0.005242 0.004162 0.006836 -0.01216 Dist.means[1] Dist.means[2] Dist.means[3] Dist.means[4] g0 gamma.copper[1] Lag 0 1.000000 1.00000 1.000000 1.000000 1.000000 NaN Lag 10 0.370254 0.23675 0.224412 0.199140 0.365289 NaN Lag 50 0.039525 0.04968 0.053600 0.037593 0.074439 NaN Lag 100 0.036857 0.01343 -0.011120 0.021074 0.028816 NaN Lag 500 -0.001278 -0.01687 0.003392 -0.007494 -0.005242 NaN gamma.copper[2] gamma.copper[3] sd.Copper sd.CopperDist sd.Dist sd.Plate sd.Resid Lag 0 1.000000 1.00000 1.00000 1.0000000 1.000000 1.000000 1.0000 Lag 10 0.338192 0.35908 0.35473 0.1967879 0.240357 0.499128 0.1724 Lag 50 0.041783 0.09064 0.09519 0.0425070 0.059514 0.098619 0.1536 Lag 100 0.015923 0.01055 0.01222 -0.0004626 0.042971 0.003585 0.1715 Lag 500 -0.004293 -0.01162 -0.00801 -0.0060531 0.006861 0.011242 0.1723 sigma.copper sigma.copperdist sigma.dist sigma.plate sigma.res Lag 0 1.000000 1.000000 1.00000 1.000000 1.000000 Lag 10 0.130953 0.199758 0.81922 0.483532 0.062755 Lag 50 0.001727 0.006590 0.46056 0.084524 0.005242 Lag 100 0.002638 -0.016696 0.31695 0.007884 -0.015005 Lag 500 0.016163 -0.005571 -0.03198 0.007846 0.014716raftery.diag(as.mcmc(copper.r2jags.a))
[[1]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[2]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[3]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
- Matrix parameterization - random intercepts model (JAGS)
Matrix parameterization JAGS code
library(coda) plot(as.mcmc(copper.r2jags.b))
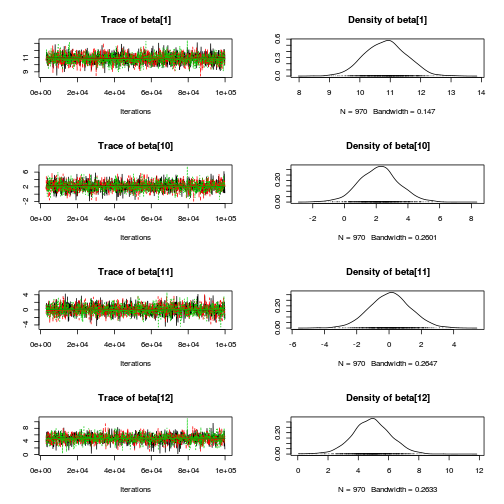
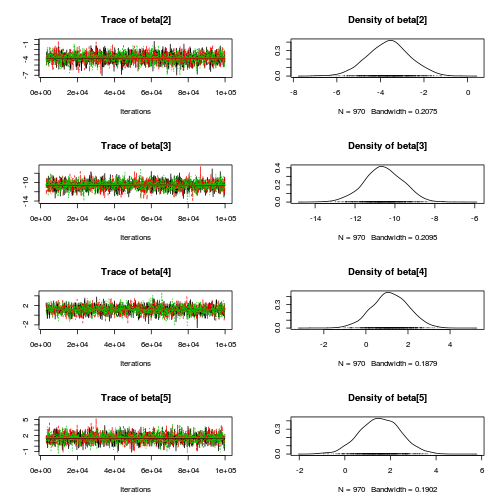
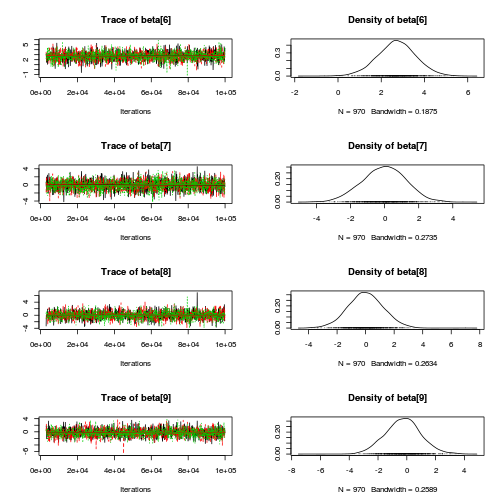
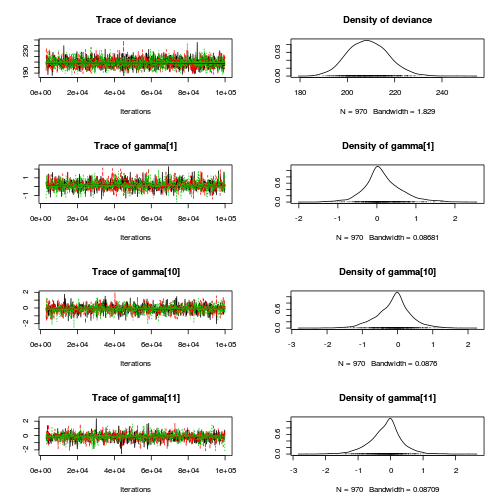
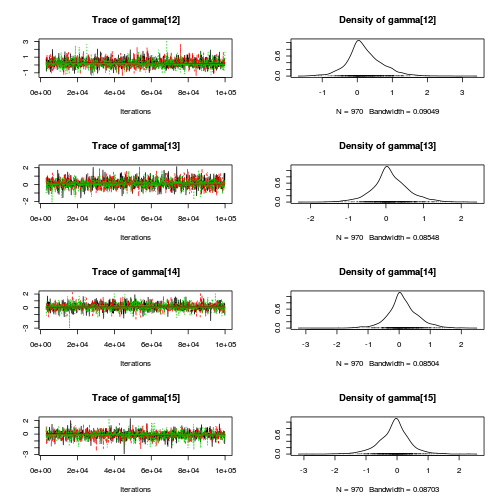
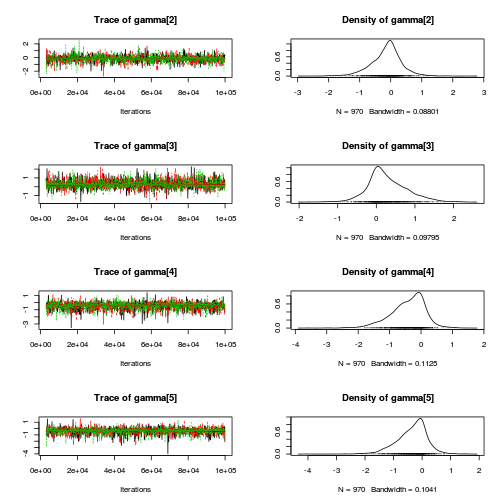
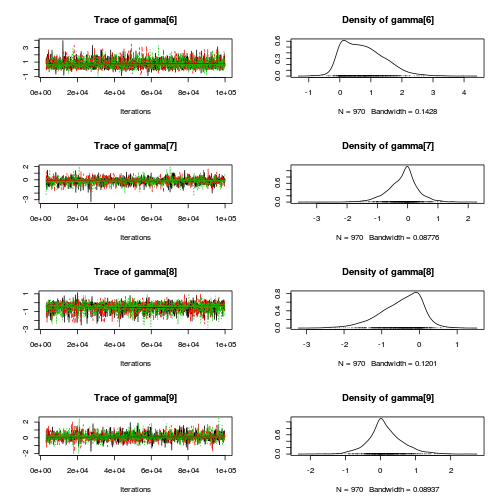
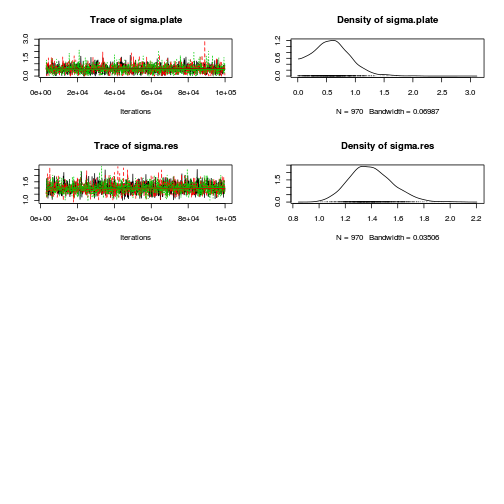
autocorr.diag(as.mcmc(copper.r2jags.b))
beta[1] beta[10] beta[11] beta[12] beta[2] beta[3] beta[4] beta[5] beta[6] Lag 0 1.000000 1.0000000 1.000000 1.000000 1.000000 1.00000 1.000000 1.000000 1.000000 Lag 100 -0.041354 -0.0252548 -0.009905 -0.036309 -0.031895 -0.03063 -0.039075 -0.016456 -0.019011 Lag 500 0.002787 -0.0340955 -0.008254 -0.028329 -0.010151 -0.01609 -0.005723 -0.005079 -0.005222 Lag 1000 -0.003429 -0.0123915 0.029828 0.019994 -0.009102 0.02454 0.015020 -0.027414 0.004205 Lag 5000 0.004199 0.0002263 -0.012197 0.003203 0.017552 0.01796 -0.011300 -0.026634 -0.005805 beta[7] beta[8] beta[9] deviance gamma[1] gamma[10] gamma[11] gamma[12] Lag 0 1.000000 1.000000 1.0000000 1.0000000 1.000000 1.0000000 1.0000000 1.000000 Lag 100 -0.035416 -0.026834 -0.0004922 0.0009833 0.042959 0.0007665 0.0007544 0.003912 Lag 500 -0.004009 -0.004309 -0.0131333 0.0092935 0.008917 0.0045661 0.0135233 -0.026427 Lag 1000 0.019981 0.022475 -0.0063841 0.0218645 0.012665 0.0011814 -0.0002279 -0.013483 Lag 5000 -0.004554 0.007378 -0.0266814 -0.0034981 0.026812 0.0353201 0.0152535 0.020084 gamma[13] gamma[14] gamma[15] gamma[2] gamma[3] gamma[4] gamma[5] gamma[6] gamma[7] Lag 0 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 1.0000000 Lag 100 -0.001690 -0.002156 -0.038614 -0.033022 0.017332 -0.012765 0.036526 0.031150 0.0033998 Lag 500 -0.011793 0.002564 0.015016 -0.008493 0.012065 0.014507 0.007995 -0.030317 0.0051025 Lag 1000 -0.005823 -0.031693 -0.001809 0.009253 0.010989 -0.003850 -0.013820 -0.014619 0.0008797 Lag 5000 0.028246 -0.008923 0.003784 0.006556 -0.003794 -0.004509 0.032079 0.007891 -0.0097967 gamma[8] gamma[9] sigma.plate sigma.res Lag 0 1.000000 1.000000 1.000000 1.000000 Lag 100 -0.018342 -0.004544 0.066395 -0.001730 Lag 500 -0.012145 -0.003570 -0.009921 -0.014436 Lag 1000 0.002001 0.001041 -0.016111 -0.005767 Lag 5000 -0.025022 0.022340 0.010262 0.021442raftery.diag(as.mcmc(copper.r2jags.b))
[[1]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[2]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[3]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
- Matrix parameterization - random intercepts model (STAN)
Matrix parameterization STAN code
library(coda) plot(copper.mcmc.c)
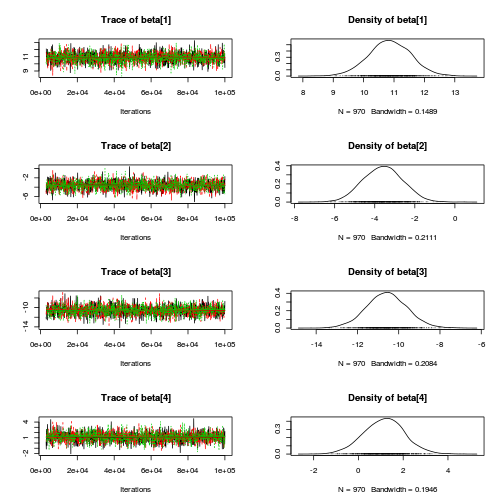
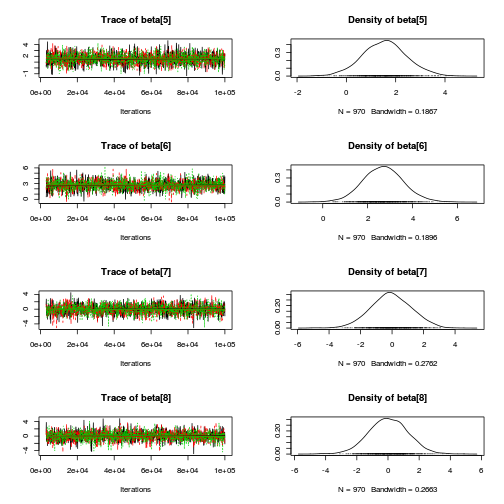
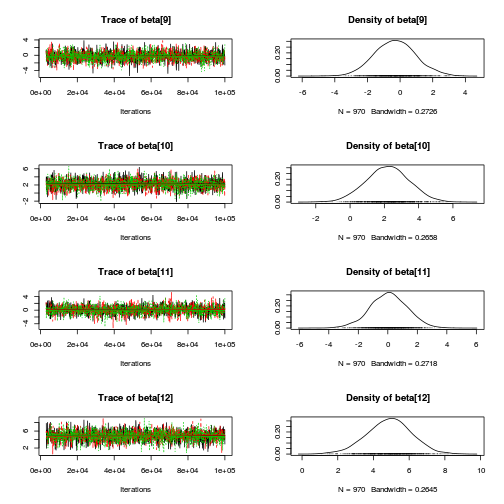
autocorr.diag(copper.mcmc.c)
beta[1] beta[2] beta[3] beta[4] beta[5] beta[6] beta[7] beta[8] beta[9] Lag 0 1.0000000 1.000000 1.0000000 1.00000 1.000000 1.00000 1.000000 1.000000 1.000000 Lag 100 -0.0041836 0.026513 -0.0288361 0.02138 0.003487 0.01788 0.011714 -0.007848 -0.005847 Lag 500 0.0045595 0.001960 0.0117408 0.01542 -0.018840 -0.01943 0.010406 0.012094 0.006645 Lag 1000 -0.0378176 0.003465 -0.0109335 -0.02404 -0.037739 -0.00797 0.007708 -0.019307 0.004713 Lag 5000 0.0004615 0.004327 0.0004766 0.01608 -0.021629 0.01313 0.020188 0.046152 -0.026899 beta[10] beta[11] beta[12] gamma[1] gamma[2] gamma[3] gamma[4] gamma[5] gamma[6] Lag 0 1.000000 1.000000 1.0000000 1.000000 1.000000 1.000000 1.000000 1.0000000 1.000000 Lag 100 -0.011190 0.003055 0.0011763 -0.006273 0.004846 -0.009932 -0.001295 0.0007406 0.007216 Lag 500 -0.036970 0.011710 -0.0138174 -0.010697 -0.008571 -0.011807 -0.020430 0.0357357 0.016466 Lag 1000 -0.019349 0.022190 0.0184706 -0.007203 -0.025243 0.003653 -0.009460 -0.0399290 0.004868 Lag 5000 0.006422 0.021870 -0.0006324 0.010839 -0.014729 -0.014651 -0.003738 0.0020064 0.015387 gamma[7] gamma[8] gamma[9] gamma[10] gamma[11] gamma[12] gamma[13] gamma[14] gamma[15] Lag 0 1.000000 1.000000 1.000000 1.000000 1.00000 1.000000 1.000000 1.000000 1.000000 Lag 100 0.001514 -0.003081 -0.034034 -0.010140 -0.01579 -0.009395 0.022341 0.002294 0.004615 Lag 500 -0.008475 -0.033721 0.019650 -0.037465 -0.01685 -0.010390 0.007332 -0.017786 -0.005211 Lag 1000 0.070823 0.008980 0.006527 -0.008765 -0.03406 -0.006356 0.045294 0.001893 -0.024810 Lag 5000 0.026074 -0.010940 -0.033948 -0.004283 -0.02907 -0.003164 0.005681 0.015060 0.022194 sigma sigma_Z lp__ Lag 0 1.000000 1.000000 1.0000000 Lag 100 0.017216 0.002634 0.0004995 Lag 500 0.021892 -0.020506 -0.0003506 Lag 1000 -0.007311 -0.003676 -0.0276602 Lag 5000 -0.025705 0.013517 0.0240491raftery.diag(copper.mcmc.c)
[[1]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[2]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[3]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
- Effects parameterization - random intercepts model (JAGS)
- Explore the parameter estimates
Full effects parameterization JAGS code
library(R2jags) print(copper.r2jags.a)
Inference for Bugs model at "../downloads/BUGSscripts/ws9.9bQ2.1a.txt", fit using jags, 3 chains, each with 16667 iterations (first 2000 discarded), n.thin = 10 n.sims = 4401 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff Copper.means[1] 10.853 0.720 9.410 10.381 10.859 11.321 12.235 1.001 3700 Copper.means[2] 6.966 0.684 5.671 6.517 6.967 7.412 8.318 1.001 3800 Copper.means[3] 0.493 0.690 -0.874 0.037 0.499 0.947 1.839 1.002 1500 CopperDist.means[1,1] 10.853 0.720 9.410 10.381 10.859 11.321 12.235 1.001 3700 CopperDist.means[2,1] 6.966 0.684 5.671 6.517 6.967 7.412 8.318 1.001 3800 CopperDist.means[3,1] 0.493 0.690 -0.874 0.037 0.499 0.947 1.839 1.002 1500 CopperDist.means[1,2] 12.008 0.676 10.668 11.563 12.018 12.459 13.309 1.001 3500 CopperDist.means[2,2] 8.405 0.674 7.055 7.963 8.406 8.854 9.687 1.001 4400 CopperDist.means[3,2] 1.579 0.679 0.255 1.121 1.584 2.026 2.908 1.001 4400 CopperDist.means[1,3] 12.428 0.642 11.152 12.018 12.414 12.838 13.708 1.001 2700 CopperDist.means[2,3] 8.591 0.683 7.245 8.135 8.586 9.052 9.951 1.001 4400 CopperDist.means[3,3] 3.932 0.681 2.586 3.485 3.920 4.380 5.298 1.001 4400 CopperDist.means[1,4] 13.523 0.708 12.190 13.032 13.531 13.989 14.896 1.001 2500 CopperDist.means[2,4] 10.088 0.676 8.750 9.641 10.078 10.546 11.434 1.001 4400 CopperDist.means[3,4] 7.543 0.720 6.117 7.056 7.557 8.034 8.912 1.001 4400 Dist.means[1] 6.107 0.366 5.367 5.869 6.115 6.340 6.827 1.002 4400 Dist.means[2] 7.262 0.798 5.725 6.724 7.267 7.800 8.811 1.001 4400 Dist.means[3] 7.682 0.779 6.113 7.172 7.691 8.187 9.228 1.001 4400 Dist.means[4] 8.777 0.826 7.159 8.223 8.754 9.324 10.417 1.001 3500 beta.CopperDist[1,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.CopperDist[2,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.CopperDist[3,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.CopperDist[1,2] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.CopperDist[2,2] 0.284 1.240 -2.158 -0.561 0.274 1.106 2.751 1.001 4400 beta.CopperDist[3,2] -0.068 1.207 -2.480 -0.874 -0.078 0.760 2.271 1.001 3700 beta.CopperDist[1,3] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.CopperDist[2,3] 0.050 1.214 -2.305 -0.756 0.051 0.841 2.413 1.001 3900 beta.CopperDist[3,3] 1.865 1.198 -0.556 1.074 1.873 2.670 4.279 1.001 3300 beta.CopperDist[1,4] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.CopperDist[2,4] 0.452 1.206 -1.862 -0.367 0.463 1.272 2.824 1.001 4100 beta.CopperDist[3,4] 4.381 1.332 1.695 3.507 4.404 5.274 6.975 1.002 1700 beta.dist[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.dist[2] 1.155 0.875 -0.570 0.573 1.158 1.737 2.857 1.001 4400 beta.dist[3] 1.575 0.874 -0.170 1.001 1.585 2.135 3.320 1.001 4400 beta.dist[4] 2.670 0.911 0.926 2.069 2.660 3.274 4.489 1.001 4400 beta.plate[1] 10.968 0.761 9.466 10.450 10.967 11.482 12.429 1.001 4400 beta.plate[2] 11.627 0.875 9.934 11.039 11.618 12.213 13.359 1.001 3400 beta.plate[3] 10.449 0.786 8.825 9.918 10.476 10.998 11.951 1.001 4400 beta.plate[4] 10.505 0.784 8.885 10.000 10.528 11.031 12.011 1.001 4400 beta.plate[5] 10.728 0.759 9.153 10.242 10.743 11.233 12.223 1.002 4400 beta.plate[6] 6.861 0.720 5.438 6.385 6.857 7.329 8.312 1.001 4400 beta.plate[7] 6.505 0.763 4.948 6.012 6.509 7.014 7.976 1.001 4400 beta.plate[8] 7.245 0.741 5.845 6.743 7.233 7.753 8.734 1.001 2900 beta.plate[9] 7.107 0.724 5.709 6.613 7.100 7.582 8.560 1.001 4400 beta.plate[10] 7.129 0.736 5.700 6.640 7.123 7.618 8.582 1.001 4400 beta.plate[11] 0.420 0.736 -1.026 -0.071 0.421 0.910 1.876 1.001 2500 beta.plate[12] 0.343 0.737 -1.091 -0.146 0.340 0.848 1.790 1.003 980 beta.plate[13] 0.387 0.737 -1.022 -0.108 0.393 0.879 1.808 1.002 1300 beta.plate[14] 0.707 0.738 -0.735 0.215 0.709 1.188 2.144 1.001 3500 beta.plate[15] 0.621 0.736 -0.835 0.135 0.623 1.109 2.066 1.002 1700 g0 10.853 0.720 9.410 10.381 10.859 11.321 12.235 1.001 3700 gamma.copper[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 gamma.copper[2] -3.886 0.986 -5.802 -4.551 -3.875 -3.221 -1.907 1.002 1800 gamma.copper[3] -10.360 0.995 -12.307 -11.007 -10.357 -9.709 -8.362 1.001 3900 sd.Copper 5.256 0.495 4.244 4.936 5.251 5.578 6.246 1.001 2800 sd.CopperDist 1.484 0.351 0.817 1.248 1.477 1.714 2.202 1.001 3500 sd.Dist 1.208 0.360 0.511 0.962 1.205 1.448 1.937 1.001 4400 sd.Plate 0.528 0.271 0.045 0.326 0.528 0.724 1.048 1.004 1400 sd.Resid 4.304 0.000 4.304 4.304 4.304 4.304 4.304 1.000 1 sigma.copper 29.628 25.472 2.924 8.959 20.406 44.181 91.539 1.001 2800 sigma.copperdist 2.660 1.611 0.912 1.690 2.263 3.160 6.733 1.001 3200 sigma.dist 5.912 12.445 0.133 0.880 1.769 4.373 47.253 1.015 300 sigma.plate 0.587 0.334 0.050 0.343 0.560 0.795 1.318 1.003 1700 sigma.res 1.408 0.166 1.121 1.289 1.398 1.511 1.761 1.002 2300 deviance 209.192 8.606 193.679 203.023 208.938 214.934 226.967 1.002 1400 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 37.0 and DIC = 246.2 DIC is an estimate of expected predictive error (lower deviance is better).Matrix parameterization JAGS codelibrary(R2jags) print(copper.r2jags.b)
Inference for Bugs model at "../downloads/BUGSscripts/ws9.4bQ2.1b.txt", fit using jags, 3 chains, each with 1e+05 iterations (first 3000 discarded), n.thin = 100 n.sims = 2910 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff beta[1] 10.849 0.684 9.531 10.377 10.857 11.304 12.152 1.002 1200 beta[2] -3.617 0.977 -5.522 -4.272 -3.608 -2.979 -1.701 1.003 740 beta[3] -10.573 0.976 -12.521 -11.215 -10.588 -9.909 -8.713 1.002 1900 beta[4] 1.161 0.877 -0.575 0.588 1.157 1.759 2.852 1.001 2900 beta[5] 1.552 0.885 -0.147 0.950 1.550 2.160 3.270 1.003 690 beta[6] 2.712 0.894 0.923 2.118 2.715 3.286 4.471 1.001 2900 beta[7] -0.041 1.274 -2.579 -0.876 -0.004 0.828 2.370 1.001 2900 beta[8] 0.007 1.241 -2.452 -0.810 -0.007 0.831 2.453 1.001 2900 beta[9] -0.264 1.237 -2.641 -1.084 -0.241 0.529 2.223 1.002 1300 beta[10] 2.175 1.244 -0.309 1.332 2.181 2.953 4.666 1.001 2900 beta[11] 0.066 1.279 -2.410 -0.760 0.080 0.889 2.605 1.001 2900 beta[12] 4.870 1.260 2.376 4.055 4.890 5.696 7.346 1.011 2900 gamma[1] 0.156 0.490 -0.774 -0.122 0.095 0.419 1.276 1.001 2900 gamma[2] -0.140 0.492 -1.214 -0.407 -0.088 0.141 0.805 1.001 2900 gamma[3] 0.285 0.511 -0.606 -0.029 0.201 0.581 1.435 1.001 2900 gamma[4] -0.423 0.547 -1.665 -0.732 -0.335 -0.030 0.469 1.001 2900 gamma[5] -0.360 0.531 -1.518 -0.655 -0.275 -0.006 0.525 1.002 1300 gamma[6] 0.765 0.664 -0.170 0.217 0.677 1.193 2.238 1.001 2900 gamma[7] -0.153 0.493 -1.219 -0.430 -0.091 0.117 0.781 1.003 850 gamma[8] -0.491 0.558 -1.786 -0.829 -0.397 -0.067 0.370 1.001 2900 gamma[9] 0.138 0.497 -0.810 -0.145 0.082 0.412 1.255 1.001 2900 gamma[10] -0.106 0.486 -1.120 -0.386 -0.061 0.160 0.877 1.002 1500 gamma[11] -0.148 0.495 -1.204 -0.415 -0.096 0.128 0.816 1.002 1100 gamma[12] 0.213 0.503 -0.710 -0.076 0.152 0.488 1.308 1.003 760 gamma[13] 0.118 0.488 -0.886 -0.145 0.079 0.388 1.178 1.002 2200 gamma[14] 0.121 0.492 -0.871 -0.145 0.081 0.385 1.185 1.001 2900 gamma[15] -0.092 0.502 -1.197 -0.359 -0.055 0.183 0.902 1.002 1800 sigma.plate 0.588 0.328 0.044 0.352 0.572 0.788 1.287 1.003 1600 sigma.res 1.397 0.164 1.102 1.284 1.386 1.502 1.745 1.001 2900 deviance 208.815 8.504 193.162 202.760 208.517 214.590 226.162 1.001 2300 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 36.2 and DIC = 245.0 DIC is an estimate of expected predictive error (lower deviance is better).Matrix parameterization STAN codelibrary(R2jags) print(copper.rstan.a)
Inference for Stan model: rstanString. 3 chains, each with iter=1e+05; warmup=3000; thin=100; post-warmup draws per chain=970, total post-warmup draws=2910. mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat beta[1] 10.86 0.01 0.69 9.49 10.40 10.86 11.34 12.21 2910 1 beta[2] -3.59 0.02 0.98 -5.49 -4.28 -3.59 -2.93 -1.70 2670 1 beta[3] -10.58 0.02 0.98 -12.51 -11.23 -10.58 -9.93 -8.63 2910 1 beta[4] 1.15 0.02 0.91 -0.66 0.54 1.16 1.75 2.95 2672 1 beta[5] 1.56 0.02 0.89 -0.22 0.96 1.56 2.12 3.35 2909 1 beta[6] 2.69 0.02 0.90 0.94 2.10 2.69 3.28 4.50 2807 1 beta[7] -0.06 0.02 1.29 -2.58 -0.92 -0.08 0.80 2.47 2670 1 beta[8] 0.03 0.02 1.27 -2.52 -0.80 0.02 0.86 2.50 2910 1 beta[9] -0.30 0.02 1.27 -2.74 -1.16 -0.31 0.54 2.24 2910 1 beta[10] 2.19 0.02 1.25 -0.26 1.37 2.20 3.03 4.60 2910 1 beta[11] 0.06 0.02 1.27 -2.49 -0.78 0.06 0.91 2.51 2892 1 beta[12] 4.88 0.02 1.25 2.32 4.06 4.92 5.70 7.26 2910 1 gamma[1] 0.15 0.01 0.49 -0.80 -0.12 0.11 0.39 1.21 2910 1 gamma[2] -0.13 0.01 0.50 -1.23 -0.40 -0.09 0.14 0.85 2880 1 gamma[3] 0.27 0.01 0.51 -0.63 -0.05 0.19 0.54 1.43 2910 1 gamma[4] -0.42 0.01 0.55 -1.66 -0.73 -0.32 -0.03 0.46 2910 1 gamma[5] -0.36 0.01 0.55 -1.56 -0.68 -0.28 0.00 0.60 2910 1 gamma[6] 0.77 0.01 0.67 -0.18 0.23 0.69 1.19 2.26 2861 1 gamma[7] -0.16 0.01 0.50 -1.29 -0.44 -0.10 0.13 0.79 2910 1 gamma[8] -0.49 0.01 0.56 -1.78 -0.83 -0.40 -0.08 0.37 2910 1 gamma[9] 0.14 0.01 0.50 -0.82 -0.14 0.10 0.41 1.20 2910 1 gamma[10] -0.12 0.01 0.50 -1.22 -0.38 -0.07 0.16 0.83 2910 1 gamma[11] -0.13 0.01 0.50 -1.25 -0.39 -0.09 0.14 0.81 2910 1 gamma[12] 0.20 0.01 0.50 -0.71 -0.10 0.14 0.49 1.33 2910 1 gamma[13] 0.12 0.01 0.50 -0.86 -0.15 0.08 0.39 1.23 2648 1 gamma[14] 0.12 0.01 0.49 -0.85 -0.14 0.08 0.37 1.21 2895 1 gamma[15] -0.08 0.01 0.51 -1.18 -0.34 -0.04 0.20 0.93 2884 1 sigma 1.40 0.00 0.16 1.13 1.29 1.39 1.50 1.75 2811 1 sigma_Z 0.59 0.01 0.33 0.07 0.35 0.57 0.79 1.32 2899 1 lp__ -45.79 0.16 8.69 -59.87 -51.27 -46.89 -41.98 -23.18 2910 1 Samples were drawn using NUTS(diag_e) at Fri Jan 30 14:27:46 2015. For each parameter, n_eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence, Rhat=1). - Figure time
Matrix parameterization JAGS code
nms <- colnames(copper.mcmc.b) pred <- grep('beta',nms) coefs <- copper.mcmc.b[,pred] newdata <- expand.grid(COPPER=levels(copper$COPPER), DIST=levels(copper$DIST)) Xmat <- model.matrix(~COPPER*DIST, newdata) pred <- coefs %*% t(Xmat) library(plyr) newdata <- cbind(newdata, adply(pred, 2, function(x) { data.frame(Median=median(x), HPDinterval(as.mcmc(x)), HPDinterval(as.mcmc(x), p=0.68)) })) newdata
COPPER DIST X1 Median lower upper lower.1 upper.1 1 control 1 1 10.8509 9.5376 12.248 10.1278 11.4944 2 Week 1 1 2 7.2591 5.8871 8.671 6.5578 7.9453 3 Week 2 1 3 0.2291 -1.1643 1.498 -0.4237 0.9722 4 control 2 4 11.9848 10.6573 13.364 11.2657 12.6401 5 Week 1 2 5 8.3605 7.0515 9.756 7.7501 9.1005 6 Week 2 2 6 1.4535 0.0694 2.797 0.7798 2.1493 7 control 3 7 12.3968 11.0182 13.717 11.7202 13.0388 8 Week 1 3 8 8.5179 7.0922 9.879 7.7176 9.0942 9 Week 2 3 9 3.9899 2.5919 5.333 3.3647 4.7204 10 control 4 10 13.5628 12.2433 14.972 12.8406 14.1892 11 Week 1 4 11 10.0036 8.7669 11.485 9.3493 10.7208 12 Week 2 4 12 7.8471 6.5727 9.332 7.1806 8.5485
library(ggplot2) p1 <-ggplot(newdata, aes(y=Median, x=as.integer(DIST), fill=COPPER)) + geom_line(aes(linetype=COPPER)) + #geom_line()+ geom_linerange(aes(ymin=lower, ymax=upper), show_guide=FALSE)+ geom_linerange(aes(ymin=lower.1, ymax=upper.1), size=2,show_guide=FALSE)+ geom_point(size=3, shape=21)+ scale_y_continuous(expression(Density~of~worms~(cm^2)))+ scale_x_continuous('Distance from plate center')+ scale_fill_manual('Treatment', breaks=c('control','Week 1', 'Week 2'), values=c('black','white','grey'), labels=c('Control','Week 1','Week 2'))+ scale_linetype_manual('Treatment', breaks=c('control','Week 1', 'Week 2'), values=c('solid','dashed','dotted'), labels=c('Control','Week 1','Week 2'))+ theme_classic() + theme(legend.justification=c(1,0), legend.position=c(1,0), legend.key.width=unit(3,'lines'), axis.title.y=element_text(vjust=2, size=rel(1.2)), axis.title.x=element_text(vjust=-1, size=rel(1.2)), plot.margin=unit(c(0.5,0.5,1,2),'lines')) copper.sd$Source <- factor(copper.sd$Source, levels=c("Resid","COPPER:DIST","DIST","PLATE","COPPER"), labels=c("Residuals", "Copper:Dist", "Dist","Plates","Copper")) copper.sd$Perc <- 100*copper.sd$Median/sum(copper.sd$Median) p2<-ggplot(copper.sd,aes(y=Source, x=Median))+ geom_vline(xintercept=0,linetype="dashed")+ geom_hline(xintercept=0)+ scale_x_continuous("Finite population \nvariance components (sd)")+ geom_errorbarh(aes(xmin=lower.1, xmax=upper.1), height=0, size=1)+ geom_errorbarh(aes(xmin=lower, xmax=upper), height=0, size=1.5)+ geom_point(size=3)+ geom_text(aes(label=sprintf("(%4.1f%%)",Perc),vjust=-1))+ theme_classic()+ theme(axis.line=element_blank(),axis.title.y=element_blank(), axis.text.y=element_text(size=12,hjust=1)) library(gridExtra) grid.arrange(p1, p2, ncol=2)
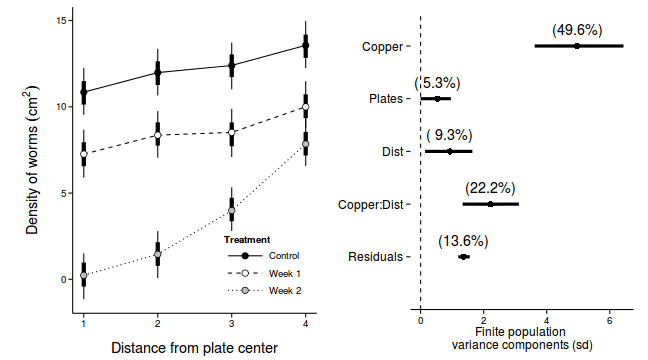
Repeated Measures
Repeated MeasuresIn an honours thesis from (1992), Mullens was investigating the ways that cane toads ( Bufo marinus ) respond to conditions of hypoxia. Toads show two different kinds of breathing patterns, lung or buccal, requiring them to be treated separately in the experiment. Her aim was to expose toads to a range of O2 concentrations, and record their breathing patterns, including parameters such as the expired volume for individual breaths. It was desirable to have around 8 replicates to compare the responses of the two breathing types, and the complication is that animals are expensive, and different individuals are likely to have different O2 profiles (leading to possibly reduced power). There are two main design options for this experiment;
- One animal per O2 treatment, 8 concentrations, 2 breathing types. With 8 replicates the experiment would require 128 animals, but that this could be analysed as a completely randomized design
- One O2 profile per animal, so that each animal would be used 8 times and only 16 animals are required (8 lung and 8 buccal breathers)
| Format of mullens.csv data file | |||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
 |
mullens <- read.table('../downloads/data/mullens.csv', header=T, sep=',', strip.white=T) head(mullens)
BREATH TOAD O2LEVEL FREQBUC SFREQBUC 1 lung a 0 10.6 3.256 2 lung a 5 18.8 4.336 3 lung a 10 17.4 4.171 4 lung a 15 16.6 4.074 5 lung a 20 9.4 3.066 6 lung a 30 11.4 3.376
Notice that the BREATH variable contains a categorical listing of the breathing type and that the first entries begin with "l" with comes after "b" in the alphabet (and therefore when BREATH is converted into a numeric, the first entry will be a 2. So we need to be careful that our codes reflect the order of the data.
Exploratory data analysis indicated that a square-root transformation of the FREQBUC (response variable) could normalize this variable and thereby address heterogeneity issues. Consequently, we too will apply a square-root transformation.
- Fit the JAGS model for the Randomized Complete Block
- Effects parameterization - random intercepts JAGS model
number of lesionsi = βSite j(i) + εi where ε ∼ N(0,σ2)
View full coderunif(1)
[1] 0.5659
modelString= ## write the model to a text file (I suggest you alter the path to somewhere more relevant ## to your system!) writeLines(modelString,con="../downloads/BUGSscripts/ws9.4bQ4.1a.txt") #sort the data set so that the breath treatments are in a more logical order mullens.sort <- mullens[order(mullens$BREATH),] mullens.sort$BREATH <- factor(mullens.sort$BREATH, levels = c("buccal", "lung")) mullens.sort$TOAD <- factor(mullens.sort$TOAD, levels=unique(mullens.sort$TOAD)) br <- with(mullens.sort,BREATH[match(unique(TOAD),TOAD)]) br <- factor(br, levels=c("buccal", "lung")) mullens.list <- with(mullens.sort, list(freqbuc=FREQBUC, o2level=as.numeric(O2LEVEL), cO2level=as.numeric(as.factor(O2LEVEL)), toad=as.numeric(TOAD), br=factor(as.numeric(br)), breath=as.numeric(BREATH), nO2level=length(levels(as.factor(O2LEVEL))), nToad=length(levels(as.factor(TOAD))), nBreath=length(levels(br)), n=nrow(mullens.sort) ) ) params <- c( "g0","gamma.breath", "beta.toad", "beta.o2level", "beta.BreathO2level", "Breath.means","O2level.means","BreathO2level.means", "sd.O2level","sd.Breath","sd.Toad","sd.Resid","sd.BreathO2level", "sigma.res","sigma.toad","sigma.breath","sigma.o2level","sigma.breatho2level" ) adaptSteps = 1000 burnInSteps = 2000 nChains = 3 numSavedSteps = 5000 thinSteps = 10 nIter = ceiling((numSavedSteps * thinSteps)/nChains) library(R2jags) ## mullens.r2jags.a <- jags(data=mullens.list, inits=NULL, # since there are three chains parameters.to.save=params, model.file="../downloads/BUGSscripts/ws9.4bQ4.1a.txt", n.chains=3, n.iter=nIter, n.burnin=burnInSteps, n.thin=thinSteps )
Compiling data graph Resolving undeclared variables Allocating nodes Initializing Reading data back into data table Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 1252 Initializing model
print(mullens.r2jags.a)
Inference for Bugs model at "../downloads/BUGSscripts/ws9.4bQ4.1a.txt", fit using jags, 3 chains, each with 16667 iterations (first 2000 discarded), n.thin = 10 n.sims = 4401 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff Breath.means[1] 4.937 0.357 4.213 4.699 4.940 5.181 5.625 1.001 3500 Breath.means[2] 1.541 0.463 0.645 1.236 1.539 1.851 2.436 1.001 4400 BreathO2level.means[1,1] 4.937 0.357 4.213 4.699 4.940 5.181 5.625 1.001 3500 BreathO2level.means[2,1] 1.541 0.463 0.645 1.236 1.539 1.851 2.436 1.001 4400 BreathO2level.means[1,2] 4.510 0.364 3.778 4.265 4.513 4.755 5.228 1.001 4400 BreathO2level.means[2,2] 2.555 0.474 1.614 2.240 2.555 2.869 3.472 1.001 4400 BreathO2level.means[1,3] 4.297 0.351 3.582 4.067 4.302 4.530 4.989 1.001 3600 BreathO2level.means[2,3] 3.367 0.460 2.463 3.056 3.372 3.671 4.255 1.001 4400 BreathO2level.means[1,4] 4.004 0.352 3.307 3.772 4.011 4.236 4.694 1.001 4400 BreathO2level.means[2,4] 3.085 0.452 2.190 2.779 3.094 3.380 3.953 1.001 4400 BreathO2level.means[1,5] 3.446 0.352 2.751 3.208 3.446 3.679 4.146 1.001 4400 BreathO2level.means[2,5] 3.225 0.462 2.325 2.912 3.210 3.531 4.132 1.001 4400 BreathO2level.means[1,6] 3.500 0.348 2.805 3.268 3.499 3.725 4.192 1.001 4400 BreathO2level.means[2,6] 3.030 0.455 2.146 2.730 3.037 3.335 3.905 1.001 4400 BreathO2level.means[1,7] 2.818 0.357 2.136 2.583 2.820 3.051 3.524 1.001 4400 BreathO2level.means[2,7] 2.817 0.460 1.900 2.509 2.819 3.117 3.739 1.001 4400 BreathO2level.means[1,8] 2.614 0.350 1.931 2.382 2.617 2.848 3.298 1.001 3300 BreathO2level.means[2,8] 2.461 0.465 1.529 2.162 2.453 2.777 3.363 1.001 4400 O2level.means[1] 3.650 0.189 3.270 3.525 3.652 3.778 4.023 1.001 4400 O2level.means[2] 3.223 0.294 2.653 3.025 3.220 3.424 3.778 1.002 1400 O2level.means[3] 3.010 0.275 2.471 2.822 3.010 3.194 3.544 1.001 4400 O2level.means[4] 2.717 0.273 2.179 2.535 2.717 2.900 3.262 1.001 3000 O2level.means[5] 2.159 0.277 1.629 1.967 2.161 2.346 2.696 1.002 2200 O2level.means[6] 2.213 0.275 1.668 2.030 2.216 2.393 2.739 1.001 4400 O2level.means[7] 1.531 0.280 0.989 1.341 1.529 1.725 2.080 1.002 1600 O2level.means[8] 1.327 0.280 0.775 1.140 1.330 1.518 1.873 1.001 2700 beta.BreathO2level[1,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.BreathO2level[2,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.BreathO2level[1,2] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.BreathO2level[2,2] 1.441 0.574 0.323 1.059 1.438 1.825 2.580 1.001 2500 beta.BreathO2level[1,3] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.BreathO2level[2,3] 2.466 0.532 1.420 2.109 2.471 2.807 3.515 1.001 4400 beta.BreathO2level[1,4] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.BreathO2level[2,4] 2.477 0.534 1.438 2.111 2.477 2.855 3.484 1.001 3400 beta.BreathO2level[1,5] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.BreathO2level[2,5] 3.176 0.533 2.130 2.812 3.179 3.543 4.211 1.002 1900 beta.BreathO2level[1,6] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.BreathO2level[2,6] 2.927 0.530 1.880 2.574 2.926 3.283 3.972 1.001 3500 beta.BreathO2level[1,7] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.BreathO2level[2,7] 3.396 0.542 2.340 3.038 3.391 3.757 4.468 1.001 3600 beta.BreathO2level[1,8] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.BreathO2level[2,8] 3.243 0.545 2.205 2.861 3.247 3.607 4.308 1.001 2400 beta.o2level[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.o2level[2] -0.427 0.355 -1.111 -0.668 -0.433 -0.187 0.278 1.002 2000 beta.o2level[3] -0.640 0.340 -1.292 -0.874 -0.646 -0.419 0.040 1.001 4400 beta.o2level[4] -0.934 0.334 -1.582 -1.159 -0.931 -0.706 -0.291 1.001 2900 beta.o2level[5] -1.492 0.337 -2.138 -1.723 -1.494 -1.267 -0.824 1.001 3100 beta.o2level[6] -1.438 0.333 -2.089 -1.663 -1.438 -1.217 -0.791 1.001 4400 beta.o2level[7] -2.120 0.339 -2.768 -2.345 -2.126 -1.893 -1.439 1.002 1600 beta.o2level[8] -2.324 0.340 -2.988 -2.557 -2.319 -2.092 -1.674 1.002 2200 beta.toad[1] 4.299 0.373 3.570 4.051 4.301 4.549 5.024 1.001 3200 beta.toad[2] 6.297 0.373 5.572 6.047 6.305 6.546 7.014 1.001 4400 beta.toad[3] 4.709 0.376 3.949 4.459 4.706 4.965 5.460 1.001 2900 beta.toad[4] 4.387 0.368 3.656 4.141 4.387 4.631 5.116 1.001 3300 beta.toad[5] 6.372 0.383 5.609 6.119 6.379 6.630 7.113 1.001 4400 beta.toad[6] 4.146 0.376 3.408 3.890 4.155 4.402 4.874 1.001 3400 beta.toad[7] 3.709 0.379 2.968 3.462 3.705 3.957 4.442 1.001 4400 beta.toad[8] 4.447 0.373 3.699 4.198 4.448 4.696 5.173 1.001 3000 beta.toad[9] 5.965 0.374 5.226 5.715 5.966 6.212 6.691 1.001 4400 beta.toad[10] 4.845 0.369 4.126 4.601 4.845 5.088 5.580 1.002 1700 beta.toad[11] 4.973 0.371 4.221 4.729 4.977 5.228 5.686 1.001 4400 beta.toad[12] 5.628 0.375 4.900 5.375 5.625 5.884 6.380 1.001 3900 beta.toad[13] 4.479 0.373 3.751 4.224 4.480 4.721 5.218 1.001 2600 beta.toad[14] 1.985 0.413 1.177 1.709 1.986 2.256 2.792 1.001 3700 beta.toad[15] 2.826 0.419 2.017 2.542 2.829 3.112 3.657 1.001 3500 beta.toad[16] 1.296 0.410 0.500 1.025 1.297 1.570 2.097 1.001 3300 beta.toad[17] 0.970 0.406 0.154 0.700 0.970 1.248 1.744 1.001 4400 beta.toad[18] 0.285 0.418 -0.532 0.004 0.289 0.560 1.084 1.001 4400 beta.toad[19] 2.189 0.413 1.375 1.909 2.194 2.465 2.993 1.001 4400 beta.toad[20] 1.590 0.418 0.772 1.310 1.587 1.870 2.406 1.001 4400 beta.toad[21] 1.259 0.411 0.451 0.992 1.251 1.539 2.061 1.001 3100 g0 4.937 0.357 4.213 4.699 4.940 5.181 5.625 1.001 3500 gamma.breath[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 gamma.breath[2] -3.396 0.589 -4.548 -3.790 -3.399 -3.007 -2.237 1.001 4400 sd.Breath 2.402 0.416 1.582 2.126 2.404 2.680 3.216 1.001 4400 sd.BreathO2level 1.483 0.206 1.080 1.344 1.486 1.621 1.889 1.002 2000 sd.O2level 0.844 0.096 0.662 0.780 0.845 0.908 1.031 1.002 2100 sd.Resid 1.483 0.000 1.483 1.483 1.483 1.483 1.483 1.000 1 sd.Toad 0.883 0.083 0.729 0.828 0.880 0.934 1.057 1.002 1400 sigma.breath 49.572 28.985 2.350 24.101 49.438 74.781 97.415 1.001 4400 sigma.breatho2level 0.979 0.517 0.366 0.663 0.875 1.166 2.224 1.001 4400 sigma.o2level 0.969 0.430 0.465 0.695 0.868 1.123 2.018 1.001 3500 sigma.res 0.881 0.056 0.779 0.842 0.878 0.916 1.000 1.001 4400 sigma.toad 0.944 0.188 0.650 0.811 0.920 1.050 1.369 1.001 4400 deviance 432.451 10.234 414.872 425.254 431.611 438.804 454.837 1.001 4400 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 52.4 and DIC = 484.8 DIC is an estimate of expected predictive error (lower deviance is better). - Matrix parameterization - random intercepts model (JAGS)
number of lesionsi = βSite j(i) + εi where ε ∼ N(0,σ2)treating Distance as a factor
View full codeWe can perform generate the finite-population standard deviation posteriors within RmodelString=" model { #Likelihood for (i in 1:n) { y[i]~dnorm(mu[i],tau.res) mu[i] <- inprod(beta[],X[i,]) + inprod(gamma[],Z[i,]) y.err[i] <- y[i] - mu[1] } #Priors and derivatives for (i in 1:nZ) { gamma[i] ~ dnorm(0,tau.toad) } for (i in 1:nX) { beta[i] ~ dnorm(0,1.0E-06) } tau.res <- pow(sigma.res,-2) sigma.res <- z/sqrt(chSq) z ~ dnorm(0, .0016)I(0,) chSq ~ dgamma(0.5, 0.5) tau.toad <- pow(sigma.toad,-2) sigma.toad <- z.toad/sqrt(chSq.toad) z.toad ~ dnorm(0, .0016)I(0,) chSq.toad ~ dgamma(0.5, 0.5) } " ## write the model to a text file (I suggest you alter the path to somewhere more relevant ## to your system!) writeLines(modelString,con="../downloads/BUGSscripts/ws9.4bQ4.1b.txt") mullens$iO2LEVEL <- mullens$O2LEVEL mullens$O2LEVEL <- factor(mullens$O2LEVEL) Xmat <- model.matrix(~BREATH*O2LEVEL, data=mullens) Zmat <- model.matrix(~-1+TOAD, data=mullens) mullens.list <- list(y=mullens$SFREQBUC, X=Xmat, nX=ncol(Xmat), Z=Zmat, nZ=ncol(Zmat), n=nrow(mullens) ) params <- c("beta","gamma", "sigma.res","sigma.toad") burnInSteps = 3000 nChains = 3 numSavedSteps = 3000 thinSteps = 100 nIter = ceiling((numSavedSteps * thinSteps)/nChains) library(R2jags) library(coda) ## mullens.r2jags.b <- jags(data=mullens.list, inits=NULL, # since there are three chains parameters.to.save=params, model.file="../downloads/BUGSscripts/ws9.4bQ4.1b.txt", n.chains=3, n.iter=nIter, n.burnin=burnInSteps, n.thin=thinSteps )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 7073 Initializing model
print(mullens.r2jags.b)
Inference for Bugs model at "../downloads/BUGSscripts/ws9.4bQ4.1b.txt", fit using jags, 3 chains, each with 1e+05 iterations (first 3000 discarded), n.thin = 100 n.sims = 2910 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff beta[1] 4.948 0.359 4.258 4.707 4.943 5.186 5.655 1.001 2900 beta[2] -3.405 0.571 -4.513 -3.784 -3.395 -3.035 -2.283 1.001 2900 beta[3] -0.219 0.346 -0.910 -0.447 -0.219 0.019 0.466 1.001 2900 beta[4] -0.549 0.349 -1.238 -0.783 -0.550 -0.315 0.143 1.001 2300 beta[5] -0.868 0.348 -1.580 -1.105 -0.866 -0.628 -0.192 1.001 2900 beta[6] -1.551 0.344 -2.226 -1.780 -1.549 -1.315 -0.886 1.001 2900 beta[7] -1.472 0.351 -2.161 -1.703 -1.472 -1.240 -0.784 1.001 2900 beta[8] -2.248 0.344 -2.920 -2.477 -2.248 -2.014 -1.585 1.001 2900 beta[9] -2.466 0.344 -3.144 -2.701 -2.473 -2.226 -1.801 1.001 2900 beta[10] 1.050 0.545 -0.044 0.698 1.046 1.404 2.137 1.001 2400 beta[11] 2.337 0.550 1.257 1.968 2.340 2.721 3.440 1.002 1400 beta[12] 2.383 0.568 1.256 2.018 2.377 2.764 3.502 1.001 2900 beta[13] 3.307 0.552 2.221 2.950 3.297 3.672 4.382 1.001 2900 beta[14] 2.994 0.564 1.886 2.631 2.995 3.368 4.078 1.001 2900 beta[15] 3.637 0.559 2.573 3.263 3.635 4.014 4.724 1.001 2900 beta[16] 3.474 0.559 2.391 3.091 3.472 3.847 4.566 1.001 2900 gamma[1] 0.428 0.426 -0.370 0.146 0.425 0.705 1.270 1.001 2500 gamma[2] -0.642 0.394 -1.442 -0.908 -0.641 -0.373 0.115 1.001 2900 gamma[3] 1.269 0.438 0.422 0.972 1.258 1.564 2.159 1.002 2600 gamma[4] 1.356 0.388 0.599 1.103 1.350 1.607 2.124 1.001 2900 gamma[5] -0.253 0.392 -1.047 -0.506 -0.252 0.009 0.510 1.001 2900 gamma[6] -0.551 0.392 -1.339 -0.814 -0.549 -0.282 0.196 1.001 2900 gamma[7] 1.419 0.399 0.640 1.149 1.418 1.685 2.202 1.001 2600 gamma[8] -0.261 0.431 -1.119 -0.552 -0.256 0.036 0.561 1.001 2400 gamma[9] -0.802 0.392 -1.554 -1.073 -0.802 -0.540 -0.031 1.001 2900 gamma[10] -0.569 0.428 -1.415 -0.854 -0.563 -0.288 0.299 1.001 2900 gamma[11] -1.253 0.391 -2.021 -1.510 -1.256 -0.984 -0.510 1.001 2200 gamma[12] -0.489 0.384 -1.253 -0.752 -0.482 -0.225 0.250 1.002 1400 gamma[13] 1.010 0.397 0.219 0.741 1.012 1.265 1.807 1.001 2000 gamma[14] -0.097 0.389 -0.868 -0.350 -0.096 0.165 0.650 1.000 2900 gamma[15] 0.029 0.379 -0.718 -0.224 0.028 0.289 0.772 1.001 2900 gamma[16] 0.677 0.381 -0.055 0.431 0.672 0.930 1.431 1.001 2900 gamma[17] -1.261 0.437 -2.112 -1.559 -1.253 -0.964 -0.409 1.001 2900 gamma[18] -0.464 0.386 -1.252 -0.713 -0.459 -0.205 0.286 1.001 2900 gamma[19] 0.642 0.430 -0.212 0.354 0.628 0.939 1.503 1.001 2900 gamma[20] 0.031 0.425 -0.817 -0.258 0.037 0.311 0.850 1.001 2900 gamma[21] -0.281 0.434 -1.139 -0.573 -0.282 -0.012 0.611 1.001 2900 sigma.res 0.878 0.055 0.778 0.839 0.876 0.911 0.994 1.001 2900 sigma.toad 0.942 0.181 0.651 0.816 0.918 1.044 1.364 1.003 980 deviance 430.909 9.736 414.255 424.034 430.244 437.014 452.334 1.001 2900 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 47.4 and DIC = 478.3 DIC is an estimate of expected predictive error (lower deviance is better).mullens.mcmc.b <- as.mcmc(mullens.r2jags.b$BUGSoutput$sims.matrix)
View full codenms <- colnames(mullens.mcmc.b) pred <- grep('beta|gamma',nms) coefs <- mullens.mcmc.b[,pred] newdata <- mullens Amat <- cbind(model.matrix(~BREATH*O2LEVEL, newdata),model.matrix(~-1+TOAD, newdata)) pred <- coefs %*% t(Amat) y.err <- t(mullens$SFREQBUC - t(pred)) a<-apply(y.err, 1,sd) resid.sd <- data.frame(Source='Resid',Median=median(a), HPDinterval(as.mcmc(a)), HPDinterval(as.mcmc(a), p=0.68)) ## Fixed effects nms <- colnames(mullens.mcmc.b) mullens.mcmc.b.beta <- mullens.mcmc.b[,grep('beta',nms)] head(mullens.mcmc.b.beta)
Markov Chain Monte Carlo (MCMC) output: Start = 1 End = 7 Thinning interval = 1 beta[1] beta[2] beta[3] beta[4] beta[5] beta[6] beta[7] beta[8] beta[9] beta[10] beta[11] [1,] 4.880 -2.496 -0.05343 -0.7481 -0.5869 -1.303 -1.188 -2.697 -2.321 0.2640 1.960 [2,] 5.575 -4.377 -0.49387 -1.1872 -1.1980 -1.767 -2.255 -2.606 -3.232 1.7431 2.340 [3,] 4.713 -1.960 0.30678 -0.2729 -0.4483 -1.553 -1.069 -2.218 -1.847 -0.4305 1.920 [4,] 5.033 -3.070 -0.14523 -0.7453 -1.2370 -1.618 -1.541 -2.823 -2.764 0.5399 2.561 [5,] 5.207 -3.984 -0.31369 -0.3434 -0.8370 -1.739 -1.573 -2.461 -2.709 1.2610 2.792 [6,] 5.140 -3.714 -0.21333 -0.5883 -0.8169 -1.614 -1.250 -2.626 -2.566 1.5681 2.016 [7,] 5.242 -4.462 -0.20462 -0.9067 -0.4907 -1.488 -1.122 -2.258 -1.868 1.8402 3.712 beta[12] beta[13] beta[14] beta[15] beta[16] [1,] 2.088 2.270 2.255 3.151 2.693 [2,] 2.700 3.605 3.430 4.048 4.091 [3,] 1.969 2.808 1.580 2.820 2.129 [4,] 2.366 3.150 2.363 3.480 3.118 [5,] 2.947 3.921 2.883 3.865 3.885 [6,] 2.056 3.228 2.589 4.363 2.790 [7,] 2.583 3.699 3.751 4.453 3.467assign <- attr(Xmat, 'assign') assign
[1] 0 1 2 2 2 2 2 2 2 3 3 3 3 3 3 3
nms <- attr(terms(model.frame(~BREATH*O2LEVEL, mullens)),'term.labels') nms
[1] "BREATH" "O2LEVEL" "BREATH:O2LEVEL"
fixed.sd <- NULL for (i in 1:max(assign)) { w <- which(i==assign) if (length(w)==1) fixed.sd<-cbind(fixed.sd, abs(mullens.mcmc.b.beta[,w])*sd(Xmat[,w])) else fixed.sd<-cbind(fixed.sd, apply(mullens.mcmc.b.beta[,w],1,sd)) } library(plyr) fixed.sd <-adply(fixed.sd,2,function(x) { data.frame(Median=median(x), HPDinterval(as.mcmc(x)), HPDinterval(as.mcmc(x), p=0.68)) }) fixed.sd<- cbind(Source=nms, fixed.sd) # Variances nms <- colnames(mullens.mcmc.b) mullens.mcmc.b.gamma <- mullens.mcmc.b[,grep('gamma',nms)] assign <- attr(Zmat, 'assign') nms <- attr(terms(model.frame(~-1+TOAD, mullens)),'term.labels') random.sd <- NULL for (i in 1:max(assign)) { w <- which(i==assign) random.sd<-cbind(random.sd, apply(mullens.mcmc.b.gamma[,w],1,sd)) } library(plyr) random.sd <-adply(random.sd,2,function(x) { data.frame(Median=median(x), HPDinterval(as.mcmc(x)), HPDinterval(as.mcmc(x), p=0.68)) }) random.sd<- cbind(Source=nms, random.sd) (mullens.sd <- rbind(subset(fixed.sd, select=-X1), subset(random.sd, select=-X1), resid.sd))
Source Median lower upper lower.1 upper.1 1 BREATH 1.6535 1.1304 2.2066 1.3990 1.9352 2 O2LEVEL 0.8735 0.6746 1.0620 0.7710 0.9664 3 BREATH:O2LEVEL 0.9666 0.6726 1.2668 0.8278 1.1344 4 TOAD 0.8775 0.7264 1.0527 0.8048 0.9575 var1 Resid 0.8679 0.8246 0.9183 0.8400 0.8875
- Matrix parameterization - random intercepts model (STAN)
View full coderstanString=" data{ int n; int nZ; int nX; vector [n] y; matrix [n,nX] X; matrix [n,nZ] Z; vector [nX] a0; matrix [nX,nX] A0; } parameters{ vector [nX] beta; real<lower=0> sigma; vector [nZ] gamma; real<lower=0> sigma_Z; } transformed parameters { vector [n] mu; mu <- X*beta + Z*gamma; } model{ // Priors beta ~ multi_normal(a0,A0); gamma ~ normal( 0 , sigma_Z ); sigma_Z ~ cauchy(0,25); sigma ~ cauchy(0,25); y ~ normal( mu , sigma ); } generated quantities { vector [n] y_err; real<lower=0> sd_Resid; y_err <- y - mu; sd_Resid <- sd(y_err); } " #sort the data set so that the copper treatments are in a more logical order library(dplyr) mullens$O2LEVEL <- factor(mullens$O2LEVEL) Xmat <- model.matrix(~BREATH*O2LEVEL, data=mullens) Zmat <- model.matrix(~-1+TOAD, data=mullens) mullens.list <- list(y=mullens$SFREQBUC, X=Xmat, nX=ncol(Xmat), Z=Zmat, nZ=ncol(Zmat), n=nrow(mullens), a0=rep(0,ncol(Xmat)), A0=diag(100000,ncol(Xmat)) ) params <- c("beta","gamma", "sigma","sigma_Z") burnInSteps = 3000 nChains = 3 numSavedSteps = 3000 thinSteps = 10 nIter = ceiling((numSavedSteps * thinSteps)/nChains) library(rstan) library(coda) ## mullens.rstan.a <- stan(data=mullens.list, model_code=rstanString, pars=c("beta","gamma","sigma","sigma_Z"), chains=nChains, iter=nIter, warmup=burnInSteps, thin=thinSteps, save_dso=TRUE )
TRANSLATING MODEL 'rstanString' FROM Stan CODE TO C++ CODE NOW. COMPILING THE C++ CODE FOR MODEL 'rstanString' NOW. SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 1). Iteration: 1 / 10000 [ 0%] (Warmup) Iteration: 1000 / 10000 [ 10%] (Warmup) Iteration: 2000 / 10000 [ 20%] (Warmup) Iteration: 3000 / 10000 [ 30%] (Warmup) Iteration: 3001 / 10000 [ 30%] (Sampling) Iteration: 4000 / 10000 [ 40%] (Sampling) Iteration: 5000 / 10000 [ 50%] (Sampling) Iteration: 6000 / 10000 [ 60%] (Sampling) Iteration: 7000 / 10000 [ 70%] (Sampling) Iteration: 8000 / 10000 [ 80%] (Sampling) Iteration: 9000 / 10000 [ 90%] (Sampling) Iteration: 10000 / 10000 [100%] (Sampling) # Elapsed Time: 4.67 seconds (Warm-up) # 13.17 seconds (Sampling) # 17.84 seconds (Total) SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 2). Iteration: 1 / 10000 [ 0%] (Warmup) Iteration: 1000 / 10000 [ 10%] (Warmup) Iteration: 2000 / 10000 [ 20%] (Warmup) Iteration: 3000 / 10000 [ 30%] (Warmup) Iteration: 3001 / 10000 [ 30%] (Sampling) Iteration: 4000 / 10000 [ 40%] (Sampling) Iteration: 5000 / 10000 [ 50%] (Sampling) Iteration: 6000 / 10000 [ 60%] (Sampling) Iteration: 7000 / 10000 [ 70%] (Sampling) Iteration: 8000 / 10000 [ 80%] (Sampling) Iteration: 9000 / 10000 [ 90%] (Sampling) Iteration: 10000 / 10000 [100%] (Sampling) # Elapsed Time: 4.56 seconds (Warm-up) # 9.77 seconds (Sampling) # 14.33 seconds (Total) SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 3). Iteration: 1 / 10000 [ 0%] (Warmup) Iteration: 1000 / 10000 [ 10%] (Warmup) Iteration: 2000 / 10000 [ 20%] (Warmup) Iteration: 3000 / 10000 [ 30%] (Warmup) Iteration: 3001 / 10000 [ 30%] (Sampling) Iteration: 4000 / 10000 [ 40%] (Sampling) Iteration: 5000 / 10000 [ 50%] (Sampling) Iteration: 6000 / 10000 [ 60%] (Sampling) Iteration: 7000 / 10000 [ 70%] (Sampling) Iteration: 8000 / 10000 [ 80%] (Sampling) Iteration: 9000 / 10000 [ 90%] (Sampling) Iteration: 10000 / 10000 [100%] (Sampling) # Elapsed Time: 4.63 seconds (Warm-up) # 15.57 seconds (Sampling) # 20.2 seconds (Total)
print(mullens.rstan.a)
Inference for Stan model: rstanString. 3 chains, each with iter=10000; warmup=3000; thin=10; post-warmup draws per chain=700, total post-warmup draws=2100. mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat beta[1] 4.94 0.01 0.36 4.23 4.70 4.94 5.18 5.62 1824 1 beta[2] -3.40 0.01 0.57 -4.50 -3.79 -3.40 -3.02 -2.27 1878 1 beta[3] -0.23 0.01 0.34 -0.89 -0.46 -0.23 -0.01 0.43 1814 1 beta[4] -0.56 0.01 0.35 -1.22 -0.79 -0.56 -0.32 0.11 1956 1 beta[5] -0.86 0.01 0.34 -1.52 -1.09 -0.87 -0.64 -0.18 1924 1 beta[6] -1.55 0.01 0.34 -2.23 -1.78 -1.55 -1.32 -0.88 2100 1 beta[7] -1.47 0.01 0.35 -2.13 -1.70 -1.46 -1.23 -0.79 2004 1 beta[8] -2.26 0.01 0.34 -2.92 -2.47 -2.25 -2.03 -1.57 2041 1 beta[9] -2.46 0.01 0.35 -3.11 -2.70 -2.46 -2.23 -1.78 1612 1 beta[10] 1.07 0.01 0.54 0.01 0.71 1.06 1.44 2.14 1856 1 beta[11] 2.35 0.01 0.55 1.28 1.97 2.35 2.74 3.46 1893 1 beta[12] 2.37 0.01 0.54 1.32 2.01 2.37 2.73 3.41 1989 1 beta[13] 3.31 0.01 0.54 2.27 2.93 3.31 3.69 4.39 2100 1 beta[14] 2.98 0.01 0.53 1.96 2.59 2.99 3.34 4.05 2100 1 beta[15] 3.64 0.01 0.55 2.54 3.25 3.63 4.01 4.70 2100 1 beta[16] 3.46 0.01 0.55 2.42 3.10 3.47 3.83 4.53 2100 1 gamma[1] 0.45 0.01 0.44 -0.39 0.16 0.45 0.74 1.29 1795 1 gamma[2] -0.63 0.01 0.40 -1.46 -0.89 -0.64 -0.38 0.17 1540 1 gamma[3] 1.28 0.01 0.45 0.43 0.99 1.28 1.57 2.16 1806 1 gamma[4] 1.37 0.01 0.39 0.63 1.11 1.36 1.63 2.16 2094 1 gamma[5] -0.22 0.01 0.39 -0.96 -0.49 -0.23 0.04 0.53 2003 1 gamma[6] -0.54 0.01 0.39 -1.30 -0.80 -0.55 -0.28 0.23 1843 1 gamma[7] 1.43 0.01 0.38 0.67 1.17 1.43 1.68 2.16 2075 1 gamma[8] -0.25 0.01 0.44 -1.10 -0.54 -0.24 0.04 0.62 2100 1 gamma[9] -0.78 0.01 0.40 -1.58 -1.04 -0.77 -0.50 -0.02 1918 1 gamma[10] -0.57 0.01 0.43 -1.44 -0.85 -0.57 -0.28 0.24 1888 1 gamma[11] -1.23 0.01 0.38 -1.99 -1.48 -1.23 -0.97 -0.48 1878 1 gamma[12] -0.48 0.01 0.38 -1.20 -0.74 -0.48 -0.21 0.26 1819 1 gamma[13] 1.02 0.01 0.40 0.27 0.76 1.02 1.29 1.81 1986 1 gamma[14] -0.08 0.01 0.39 -0.84 -0.34 -0.07 0.17 0.64 1883 1 gamma[15] 0.03 0.01 0.38 -0.72 -0.23 0.03 0.29 0.82 1870 1 gamma[16] 0.68 0.01 0.39 -0.10 0.42 0.67 0.94 1.43 1861 1 gamma[17] -1.25 0.01 0.45 -2.15 -1.54 -1.24 -0.95 -0.38 1987 1 gamma[18] -0.45 0.01 0.39 -1.23 -0.69 -0.45 -0.19 0.29 1694 1 gamma[19] 0.65 0.01 0.45 -0.19 0.36 0.65 0.95 1.55 2011 1 gamma[20] 0.06 0.01 0.45 -0.81 -0.23 0.06 0.34 0.95 2042 1 gamma[21] -0.28 0.01 0.43 -1.11 -0.56 -0.28 0.00 0.58 1975 1 sigma 0.88 0.00 0.05 0.78 0.84 0.88 0.91 1.00 2100 1 sigma_Z 0.94 0.00 0.19 0.65 0.81 0.91 1.05 1.40 2100 1 lp__ -69.80 0.11 4.97 -80.69 -72.81 -69.40 -66.24 -61.43 2080 1 Samples were drawn using NUTS(diag_e) at Sat Jan 31 08:07:18 2015. For each parameter, n_eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence, Rhat=1).mullens.mcmc.c <- rstan:::as.mcmc.list.stanfit(mullens.rstan.a) mullens.mcmc.df.c <- as.data.frame(extract(mullens.rstan.a))
- Effects parameterization - random intercepts JAGS model
- Before fully exploring the parameters, it is prudent to examine the convergence and mixing diagnostics.
Chose either any of the parameterizations (they should yield much the same) - although it is sometimes useful to explore the
different performances of effects vs matrix and JAGS vs STAN.
- Effects parameterization - random intercepts model (JAGS)
Full effects parameterization JAGS code
library(coda) plot(as.mcmc(mullens.r2jags.a))


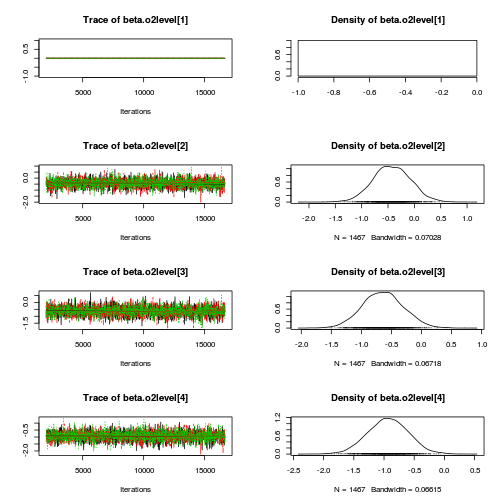
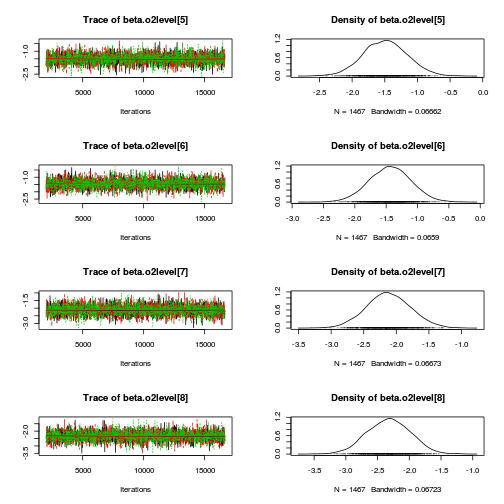
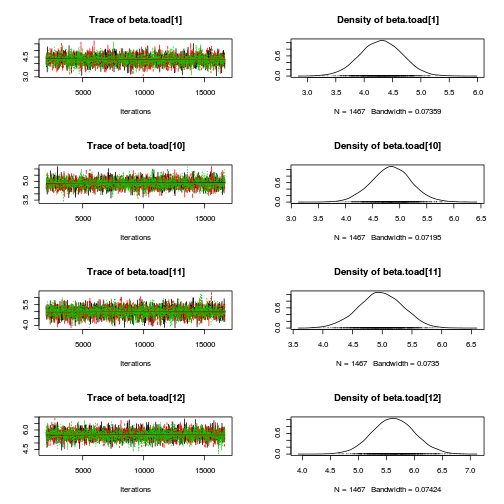
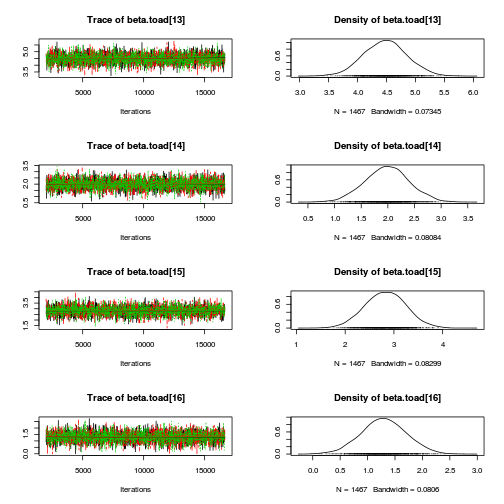
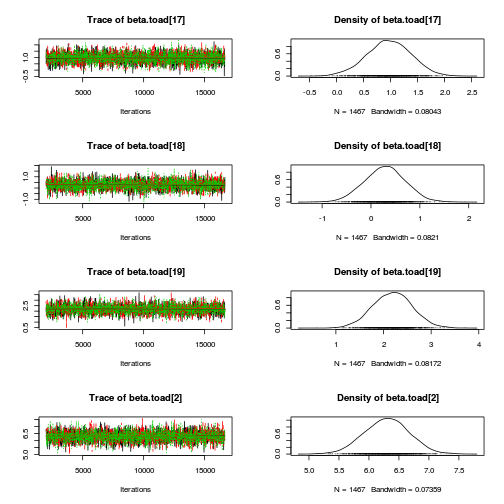
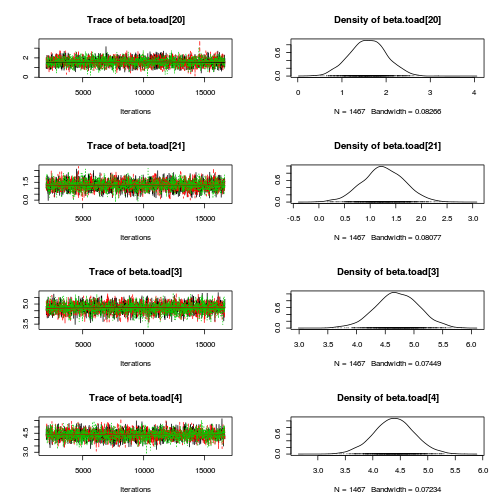
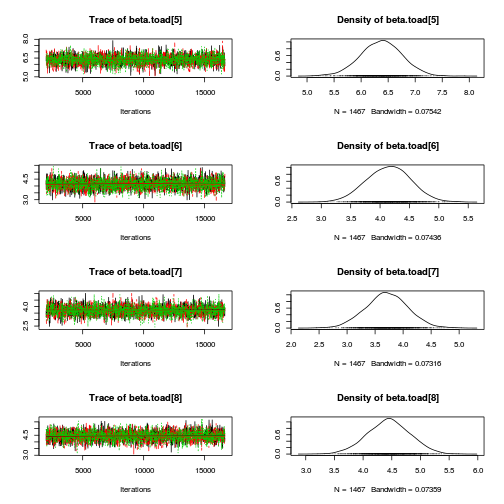
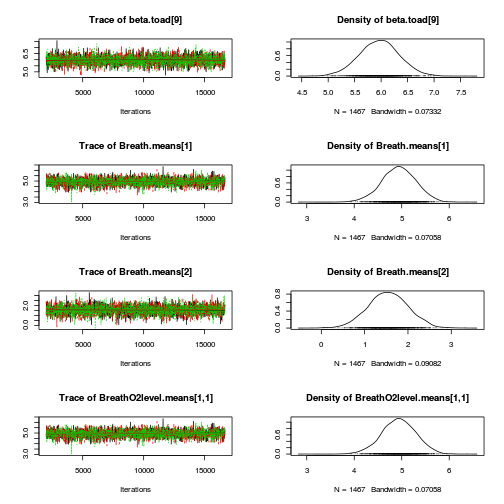
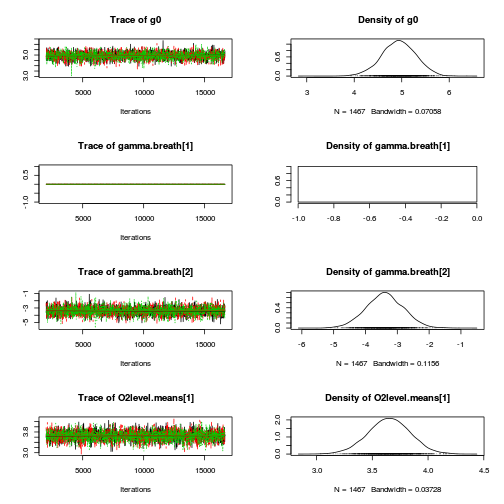
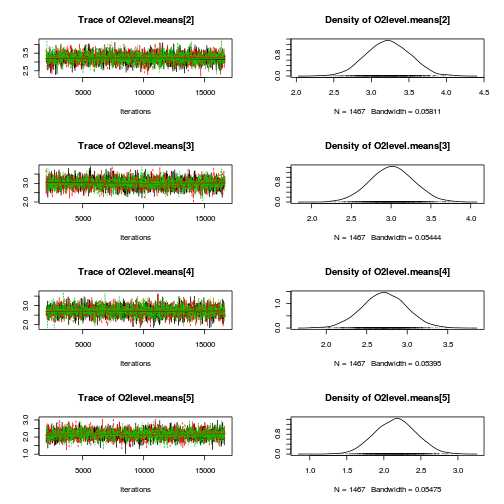
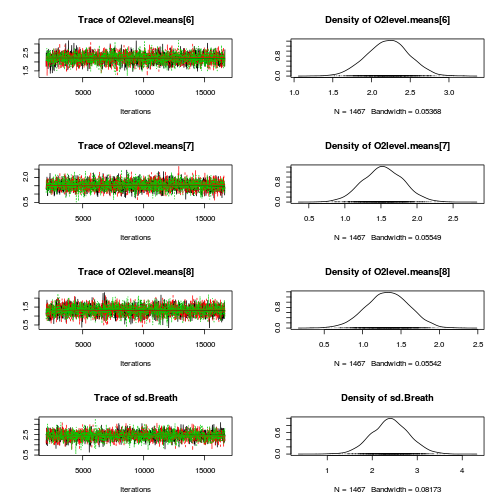
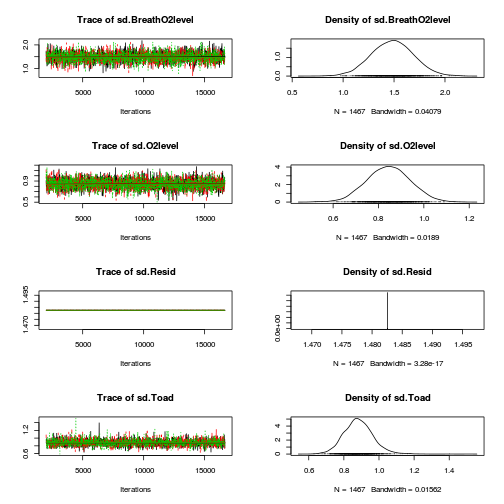
autocorr.diag(as.mcmc(mullens.r2jags.a))
beta.BreathO2level[1,1] beta.BreathO2level[1,2] beta.BreathO2level[1,3] Lag 0 NaN NaN NaN Lag 10 NaN NaN NaN Lag 50 NaN NaN NaN Lag 100 NaN NaN NaN Lag 500 NaN NaN NaN beta.BreathO2level[1,4] beta.BreathO2level[1,5] beta.BreathO2level[1,6] Lag 0 NaN NaN NaN Lag 10 NaN NaN NaN Lag 50 NaN NaN NaN Lag 100 NaN NaN NaN Lag 500 NaN NaN NaN beta.BreathO2level[1,7] beta.BreathO2level[1,8] beta.BreathO2level[2,1] Lag 0 NaN NaN NaN Lag 10 NaN NaN NaN Lag 50 NaN NaN NaN Lag 100 NaN NaN NaN Lag 500 NaN NaN NaN beta.BreathO2level[2,2] beta.BreathO2level[2,3] beta.BreathO2level[2,4] Lag 0 1.000000 1.0000000 1.000000 Lag 10 0.047119 0.0506250 0.034635 Lag 50 -0.009087 -0.0149736 -0.012821 Lag 100 0.024438 0.0015708 -0.003077 Lag 500 0.014037 0.0008682 0.002473 beta.BreathO2level[2,5] beta.BreathO2level[2,6] beta.BreathO2level[2,7] Lag 0 1.000000 1.000000 1.000000 Lag 10 0.033593 0.030292 0.005748 Lag 50 -0.015182 -0.001415 -0.013241 Lag 100 0.002849 -0.016698 0.005768 Lag 500 0.007223 0.032042 -0.004683 beta.BreathO2level[2,8] beta.o2level[1] beta.o2level[2] beta.o2level[3] beta.o2level[4] Lag 0 1.000000 NaN 1.000000 1.000000 1.000000 Lag 10 0.042863 NaN 0.027329 0.025569 0.010036 Lag 50 -0.019776 NaN -0.005901 -0.008194 0.005864 Lag 100 -0.005399 NaN 0.008269 -0.023168 -0.005180 Lag 500 -0.012152 NaN -0.001524 -0.027353 0.003771 beta.o2level[5] beta.o2level[6] beta.o2level[7] beta.o2level[8] beta.toad[1] beta.toad[10] Lag 0 1.000000 1.0000000 1.000000 1.000000 1.000000 1.00000 Lag 10 0.036928 0.0185535 0.003299 0.028334 -0.001593 0.04462 Lag 50 -0.004048 0.0182058 -0.003061 0.006838 -0.002019 0.01890 Lag 100 -0.016865 0.0005485 -0.006426 -0.008995 -0.009017 -0.03212 Lag 500 -0.010894 0.0238321 -0.016793 -0.015019 0.007171 0.02463 beta.toad[11] beta.toad[12] beta.toad[13] beta.toad[14] beta.toad[15] beta.toad[16] Lag 0 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 Lag 10 0.005842 0.006679 0.003305 0.015319 0.025592 0.026067 Lag 50 -0.019360 -0.021964 0.004889 0.006421 -0.005712 -0.027088 Lag 100 -0.013033 -0.022123 -0.004888 0.020450 0.013593 0.029467 Lag 500 0.012187 0.003450 -0.015666 0.001582 0.009586 -0.002531 beta.toad[17] beta.toad[18] beta.toad[19] beta.toad[2] beta.toad[20] beta.toad[21] Lag 0 1.0000000 1.00000 1.000000 1.000000 1.000000 1.000000 Lag 10 0.0111070 0.00820 0.008287 0.001731 0.009009 0.015540 Lag 50 -0.0227707 -0.02585 -0.026784 0.004356 -0.003178 -0.013557 Lag 100 0.0004194 0.01487 0.023727 -0.034820 0.026590 0.018269 Lag 500 0.0017732 0.01014 -0.011858 0.021306 0.030587 -0.001805 beta.toad[3] beta.toad[4] beta.toad[5] beta.toad[6] beta.toad[7] beta.toad[8] beta.toad[9] Lag 0 1.000000 1.000000 1.000000 1.0000000 1.000000 1.000000 1.0000000 Lag 10 0.026248 0.033520 -0.002853 0.0079326 0.048105 0.014745 0.0070251 Lag 50 -0.006224 -0.018632 0.011086 0.0111348 -0.008393 -0.009974 -0.0037923 Lag 100 -0.017695 0.007070 0.009695 0.0214383 0.002776 0.005192 -0.0038539 Lag 500 -0.001430 0.003278 -0.010758 -0.0003823 -0.010444 -0.003600 -0.0001833 Breath.means[1] Breath.means[2] BreathO2level.means[1,1] BreathO2level.means[1,2] Lag 0 1.000000 1.000000 1.000000 1.0000000 Lag 10 0.036582 0.005327 0.036582 -0.0007928 Lag 50 0.019244 -0.023967 0.019244 -0.0002232 Lag 100 -0.026899 -0.013783 -0.026899 -0.0243188 Lag 500 0.009972 0.017985 0.009972 -0.0250686 BreathO2level.means[1,3] BreathO2level.means[1,4] BreathO2level.means[1,5] Lag 0 1.000000 1.000000 1.000e+00 Lag 10 -0.002974 -0.008895 1.080e-02 Lag 50 -0.017006 -0.003659 1.012e-03 Lag 100 -0.011183 -0.011478 8.337e-05 Lag 500 -0.041421 -0.011624 -5.894e-03 BreathO2level.means[1,6] BreathO2level.means[1,7] BreathO2level.means[1,8] Lag 0 1.000000 1.000000 1.000000 Lag 10 -0.010967 -0.004226 0.012048 Lag 50 0.019231 0.003087 0.006620 Lag 100 -0.008105 -0.006261 0.002103 Lag 500 0.005958 -0.015027 -0.001489 BreathO2level.means[2,1] BreathO2level.means[2,2] BreathO2level.means[2,3] Lag 0 1.000000 1.000000 1.000000 Lag 10 0.005327 0.004105 -0.023403 Lag 50 -0.023967 -0.003962 -0.001983 Lag 100 -0.013783 0.007083 0.005475 Lag 500 0.017985 0.017439 0.026444 BreathO2level.means[2,4] BreathO2level.means[2,5] BreathO2level.means[2,6] Lag 0 1.0000000 1.000000 1.000000 Lag 10 0.0024557 0.006148 -0.022192 Lag 50 0.0024848 -0.003669 0.001344 Lag 100 0.0006627 0.001880 -0.002367 Lag 500 -0.0074827 0.003152 0.025256 BreathO2level.means[2,7] BreathO2level.means[2,8] deviance g0 gamma.breath[1] Lag 0 1.0000000 1.000000 1.00000 1.000000 NaN Lag 10 0.0052862 0.014475 -0.01540 0.036582 NaN Lag 50 0.0015778 -0.004027 0.02459 0.019244 NaN Lag 100 0.0048619 -0.028792 0.01528 -0.026899 NaN Lag 500 0.0008909 -0.019027 0.02504 0.009972 NaN gamma.breath[2] O2level.means[1] O2level.means[2] O2level.means[3] O2level.means[4] Lag 0 1.000000 1.000000 1.000000 1.000000 1.000000 Lag 10 0.020644 0.020517 0.022473 0.018342 -0.001684 Lag 50 0.003897 -0.016476 0.001874 -0.012485 -0.002128 Lag 100 -0.036547 0.013658 0.026012 -0.004398 0.003771 Lag 500 0.008770 0.004398 0.009155 -0.024276 -0.009611 O2level.means[5] O2level.means[6] O2level.means[7] O2level.means[8] sd.Breath Lag 0 1.000000 1.000000 1.000000 1.000000 1.000000 Lag 10 0.021182 0.008326 -0.011953 0.027630 0.020644 Lag 50 -0.005484 0.008244 -0.001709 0.011116 0.003897 Lag 100 -0.008919 -0.005162 0.008085 0.005539 -0.036547 Lag 500 -0.007931 0.031386 -0.009182 -0.027264 0.008770 sd.BreathO2level sd.O2level sd.Resid sd.Toad sigma.breath sigma.breatho2level Lag 0 1.000000 1.000000 1.0000 1.000000 1.000000 1.000000 Lag 10 0.044683 0.023225 0.2341 0.003958 0.037212 0.175480 Lag 50 -0.017903 0.004359 0.2440 0.016063 0.022793 0.005456 Lag 100 -0.002324 -0.008736 0.2556 -0.008825 -0.007311 -0.019007 Lag 500 0.006114 -0.005205 0.2381 0.002690 0.010779 -0.002027 sigma.o2level sigma.res sigma.toad Lag 0 1.000000 1.0000000 1.0000000 Lag 10 0.083060 0.0002203 -0.0109140 Lag 50 0.007404 -0.0080565 0.0241421 Lag 100 0.005133 0.0241417 -0.0097403 Lag 500 0.016259 0.0070571 -0.0002376raftery.diag(as.mcmc(mullens.r2jags.a))
[[1]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[2]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[3]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
- Matrix parameterization - random intercepts model (JAGS)
Matrix parameterization JAGS code
library(coda) plot(as.mcmc(mullens.r2jags.b))
autocorr.diag(as.mcmc(mullens.r2jags.b))
beta[1] beta[10] beta[11] beta[12] beta[13] beta[14] beta[15] beta[16] beta[2] Lag 0 1.000000 1.000000 1.00000 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 Lag 100 -0.009529 0.020223 -0.01507 -0.005496 0.007946 0.008634 -0.022852 0.019268 0.002062 Lag 500 -0.021294 -0.001715 -0.02737 -0.002556 -0.031182 0.032546 0.014163 0.005448 -0.016038 Lag 1000 -0.014413 -0.006683 -0.01883 -0.015051 0.011445 0.023777 -0.006589 -0.015876 -0.026373 Lag 5000 0.015900 -0.006645 -0.00128 0.021914 -0.005969 -0.018341 -0.019482 0.003436 0.014180 beta[3] beta[4] beta[5] beta[6] beta[7] beta[8] beta[9] deviance gamma[1] Lag 0 1.0000000 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 Lag 100 0.0177713 0.014628 0.023477 0.011826 0.011188 -0.006122 0.039054 -0.007053 0.008715 Lag 500 0.0005351 -0.016122 -0.003339 -0.047636 0.002797 -0.009193 0.016943 0.005741 -0.015718 Lag 1000 0.0103756 0.001527 0.004306 0.038051 0.021410 0.017733 0.002051 0.006209 -0.023921 Lag 5000 -0.0061523 -0.002581 -0.008062 -0.003869 0.012019 0.001681 0.017883 0.029000 0.010628 gamma[10] gamma[11] gamma[12] gamma[13] gamma[14] gamma[15] gamma[16] gamma[17] gamma[18] Lag 0 1.000000 1.00000 1.000000 1.000000 1.000000 1.000000 1.000000 1.0000000 1.000000 Lag 100 0.005849 -0.01002 -0.003408 -0.005112 -0.010012 0.003537 -0.029072 0.0004839 -0.027524 Lag 500 -0.006139 0.02423 -0.003666 -0.015927 -0.008195 0.011496 -0.014997 0.0003654 -0.018112 Lag 1000 -0.005874 -0.02346 0.012999 -0.008720 0.010156 -0.030790 0.004532 -0.0124708 -0.012201 Lag 5000 0.020177 0.03262 -0.011651 0.001316 -0.006537 -0.001419 -0.011827 -0.0303006 -0.006169 gamma[19] gamma[2] gamma[20] gamma[21] gamma[3] gamma[4] gamma[5] gamma[6] gamma[7] Lag 0 1.000000 1.000000 1.0000000 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 Lag 100 -0.031045 -0.025773 -0.0007922 0.001488 -0.007735 -0.047942 -0.003091 -0.002262 -0.013798 Lag 500 -0.033347 -0.013008 -0.0282856 -0.013552 -0.032657 -0.012294 0.014231 -0.006998 0.015697 Lag 1000 -0.013012 -0.013592 -0.0015750 -0.020833 -0.032480 -0.004777 -0.021094 -0.007831 -0.016426 Lag 5000 -0.001349 0.008919 0.0141712 0.028123 0.031898 0.013191 0.003711 -0.013612 0.002735 gamma[8] gamma[9] sigma.res sigma.toad Lag 0 1.000000 1.000000 1.000000 1.000000 Lag 100 0.014759 -0.003898 0.013504 0.018135 Lag 500 -0.015630 -0.012040 0.006071 -0.034862 Lag 1000 -0.012948 -0.028001 -0.021278 -0.008229 Lag 5000 0.003267 0.019840 0.028059 -0.002177raftery.diag(as.mcmc(mullens.r2jags.b))
[[1]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[2]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[3]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
- Matrix parameterization - random intercepts model (STAN)
Matrix parameterization STAN code
library(coda) plot(mullens.mcmc.c)
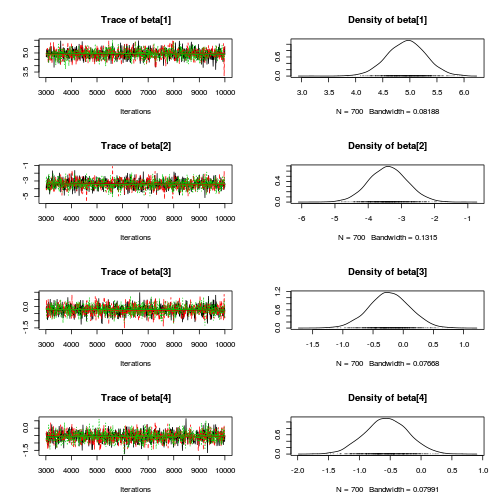
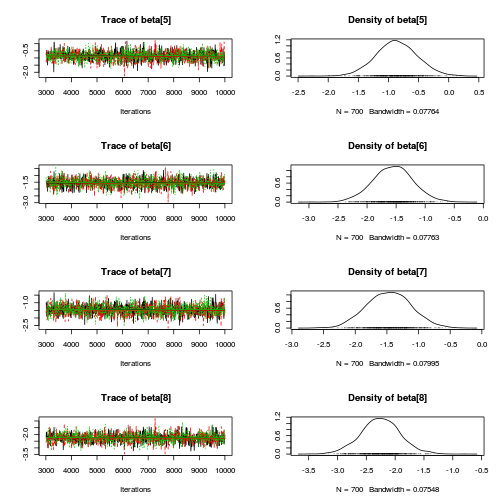
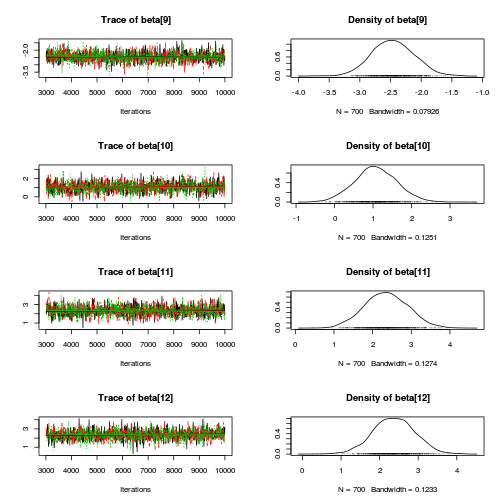
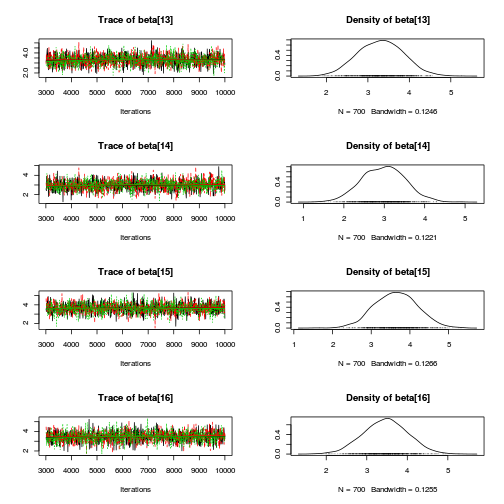
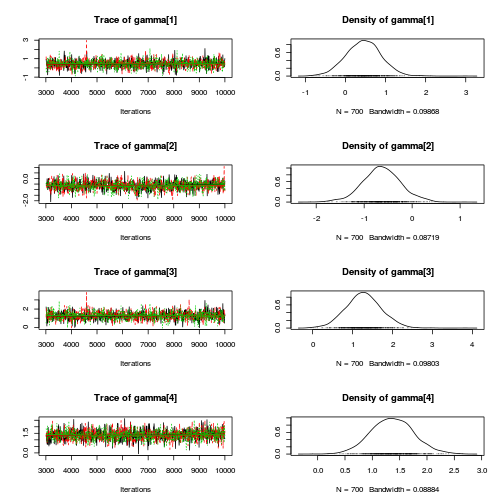
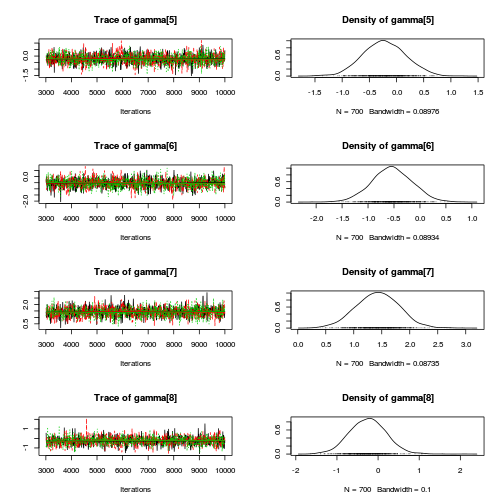
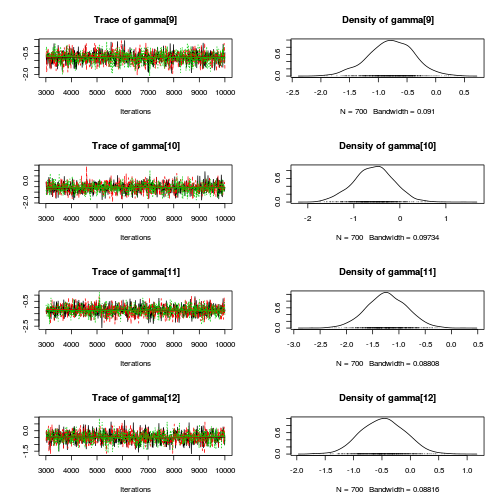
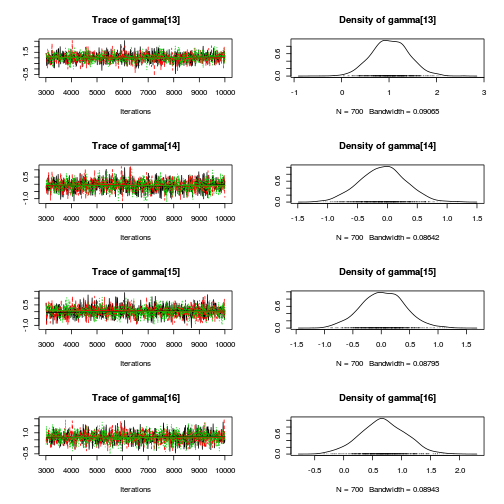
autocorr.diag(mullens.mcmc.c)
beta[1] beta[2] beta[3] beta[4] beta[5] beta[6] beta[7] beta[8] beta[9] Lag 0 1.000000 1.00000 1.000000 1.00000 1.000000 1.00000 1.000000 1.000000 1.000000 Lag 10 0.075323 0.05882 0.038002 0.02209 0.030796 -0.01069 0.019391 0.013870 0.022758 Lag 50 -0.013816 -0.04673 -0.009117 -0.01837 0.006764 -0.03159 -0.006329 -0.003814 0.003805 Lag 100 0.007330 0.02789 -0.013747 -0.01304 0.017891 -0.01109 -0.033803 0.006073 -0.013934 Lag 500 0.002964 -0.01910 -0.009464 0.03431 0.046690 -0.02060 0.009052 0.036231 -0.015621 beta[10] beta[11] beta[12] beta[13] beta[14] beta[15] beta[16] gamma[1] gamma[2] Lag 0 1.00000 1.000000 1.00000 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 Lag 10 0.02094 0.021178 0.01595 -0.014799 -0.011231 -0.007213 -0.017970 0.003187 0.041076 Lag 50 -0.01304 -0.005547 0.01283 -0.042134 -0.021393 -0.023010 -0.004467 -0.008687 0.016969 Lag 100 0.03751 -0.012920 -0.01418 0.002542 0.012572 0.012367 0.002395 -0.018293 -0.009459 Lag 500 -0.02503 0.015152 0.01160 0.017864 -0.009359 0.044098 -0.002677 -0.043487 0.010978 gamma[3] gamma[4] gamma[5] gamma[6] gamma[7] gamma[8] gamma[9] gamma[10] gamma[11] Lag 0 1.000000 1.000000 1.000000 1.0000000 1.000000 1.000000 1.000000 1.000000 1.000000 Lag 10 0.031684 0.003337 0.014156 0.0309748 0.004975 -0.001067 0.047593 0.025183 0.048205 Lag 50 -0.009988 0.002532 0.022725 -0.0123910 0.009996 -0.063657 -0.006269 -0.022206 0.023040 Lag 100 -0.010721 0.005699 -0.008436 0.0054938 0.013723 0.013245 0.016127 0.004204 -0.003023 Lag 500 -0.029727 0.017845 -0.011151 -0.0007758 0.029995 -0.018522 0.012526 0.002633 0.024268 gamma[12] gamma[13] gamma[14] gamma[15] gamma[16] gamma[17] gamma[18] gamma[19] gamma[20] Lag 0 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 Lag 10 0.066862 0.019748 0.058467 0.028520 0.065383 0.028599 0.066035 0.017727 0.012536 Lag 50 -0.011024 -0.010280 0.001222 -0.018740 0.009927 0.009400 0.006587 0.004028 -0.003746 Lag 100 -0.002287 0.048915 -0.020575 0.027193 -0.014911 -0.005238 -0.027403 -0.005643 -0.031967 Lag 500 0.002627 -0.001318 0.023401 0.004948 -0.023742 -0.009744 0.020779 -0.026538 -0.004190 gamma[21] sigma sigma_Z lp__ Lag 0 1.00000 1.00000 1.000000 1.000000 Lag 10 0.02849 -0.01214 -0.013233 0.001738 Lag 50 -0.01725 -0.03108 0.009673 -0.015617 Lag 100 -0.04150 -0.02959 -0.015358 -0.024548 Lag 500 -0.02824 0.01531 0.018352 -0.004852raftery.diag(mullens.mcmc.c)
[[1]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[2]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[3]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
- Effects parameterization - random intercepts model (JAGS)
- Explore the parameter estimates
Full effects parameterization JAGS code
library(R2jags) print(mullens.r2jags.a)
Inference for Bugs model at "../downloads/BUGSscripts/ws9.4bQ4.1a.txt", fit using jags, 3 chains, each with 16667 iterations (first 2000 discarded), n.thin = 10 n.sims = 4401 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff Breath.means[1] 4.937 0.357 4.213 4.699 4.940 5.181 5.625 1.001 3500 Breath.means[2] 1.541 0.463 0.645 1.236 1.539 1.851 2.436 1.001 4400 BreathO2level.means[1,1] 4.937 0.357 4.213 4.699 4.940 5.181 5.625 1.001 3500 BreathO2level.means[2,1] 1.541 0.463 0.645 1.236 1.539 1.851 2.436 1.001 4400 BreathO2level.means[1,2] 4.510 0.364 3.778 4.265 4.513 4.755 5.228 1.001 4400 BreathO2level.means[2,2] 2.555 0.474 1.614 2.240 2.555 2.869 3.472 1.001 4400 BreathO2level.means[1,3] 4.297 0.351 3.582 4.067 4.302 4.530 4.989 1.001 3600 BreathO2level.means[2,3] 3.367 0.460 2.463 3.056 3.372 3.671 4.255 1.001 4400 BreathO2level.means[1,4] 4.004 0.352 3.307 3.772 4.011 4.236 4.694 1.001 4400 BreathO2level.means[2,4] 3.085 0.452 2.190 2.779 3.094 3.380 3.953 1.001 4400 BreathO2level.means[1,5] 3.446 0.352 2.751 3.208 3.446 3.679 4.146 1.001 4400 BreathO2level.means[2,5] 3.225 0.462 2.325 2.912 3.210 3.531 4.132 1.001 4400 BreathO2level.means[1,6] 3.500 0.348 2.805 3.268 3.499 3.725 4.192 1.001 4400 BreathO2level.means[2,6] 3.030 0.455 2.146 2.730 3.037 3.335 3.905 1.001 4400 BreathO2level.means[1,7] 2.818 0.357 2.136 2.583 2.820 3.051 3.524 1.001 4400 BreathO2level.means[2,7] 2.817 0.460 1.900 2.509 2.819 3.117 3.739 1.001 4400 BreathO2level.means[1,8] 2.614 0.350 1.931 2.382 2.617 2.848 3.298 1.001 3300 BreathO2level.means[2,8] 2.461 0.465 1.529 2.162 2.453 2.777 3.363 1.001 4400 O2level.means[1] 3.650 0.189 3.270 3.525 3.652 3.778 4.023 1.001 4400 O2level.means[2] 3.223 0.294 2.653 3.025 3.220 3.424 3.778 1.002 1400 O2level.means[3] 3.010 0.275 2.471 2.822 3.010 3.194 3.544 1.001 4400 O2level.means[4] 2.717 0.273 2.179 2.535 2.717 2.900 3.262 1.001 3000 O2level.means[5] 2.159 0.277 1.629 1.967 2.161 2.346 2.696 1.002 2200 O2level.means[6] 2.213 0.275 1.668 2.030 2.216 2.393 2.739 1.001 4400 O2level.means[7] 1.531 0.280 0.989 1.341 1.529 1.725 2.080 1.002 1600 O2level.means[8] 1.327 0.280 0.775 1.140 1.330 1.518 1.873 1.001 2700 beta.BreathO2level[1,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.BreathO2level[2,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.BreathO2level[1,2] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.BreathO2level[2,2] 1.441 0.574 0.323 1.059 1.438 1.825 2.580 1.001 2500 beta.BreathO2level[1,3] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.BreathO2level[2,3] 2.466 0.532 1.420 2.109 2.471 2.807 3.515 1.001 4400 beta.BreathO2level[1,4] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.BreathO2level[2,4] 2.477 0.534 1.438 2.111 2.477 2.855 3.484 1.001 3400 beta.BreathO2level[1,5] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.BreathO2level[2,5] 3.176 0.533 2.130 2.812 3.179 3.543 4.211 1.002 1900 beta.BreathO2level[1,6] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.BreathO2level[2,6] 2.927 0.530 1.880 2.574 2.926 3.283 3.972 1.001 3500 beta.BreathO2level[1,7] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.BreathO2level[2,7] 3.396 0.542 2.340 3.038 3.391 3.757 4.468 1.001 3600 beta.BreathO2level[1,8] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.BreathO2level[2,8] 3.243 0.545 2.205 2.861 3.247 3.607 4.308 1.001 2400 beta.o2level[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.o2level[2] -0.427 0.355 -1.111 -0.668 -0.433 -0.187 0.278 1.002 2000 beta.o2level[3] -0.640 0.340 -1.292 -0.874 -0.646 -0.419 0.040 1.001 4400 beta.o2level[4] -0.934 0.334 -1.582 -1.159 -0.931 -0.706 -0.291 1.001 2900 beta.o2level[5] -1.492 0.337 -2.138 -1.723 -1.494 -1.267 -0.824 1.001 3100 beta.o2level[6] -1.438 0.333 -2.089 -1.663 -1.438 -1.217 -0.791 1.001 4400 beta.o2level[7] -2.120 0.339 -2.768 -2.345 -2.126 -1.893 -1.439 1.002 1600 beta.o2level[8] -2.324 0.340 -2.988 -2.557 -2.319 -2.092 -1.674 1.002 2200 beta.toad[1] 4.299 0.373 3.570 4.051 4.301 4.549 5.024 1.001 3200 beta.toad[2] 6.297 0.373 5.572 6.047 6.305 6.546 7.014 1.001 4400 beta.toad[3] 4.709 0.376 3.949 4.459 4.706 4.965 5.460 1.001 2900 beta.toad[4] 4.387 0.368 3.656 4.141 4.387 4.631 5.116 1.001 3300 beta.toad[5] 6.372 0.383 5.609 6.119 6.379 6.630 7.113 1.001 4400 beta.toad[6] 4.146 0.376 3.408 3.890 4.155 4.402 4.874 1.001 3400 beta.toad[7] 3.709 0.379 2.968 3.462 3.705 3.957 4.442 1.001 4400 beta.toad[8] 4.447 0.373 3.699 4.198 4.448 4.696 5.173 1.001 3000 beta.toad[9] 5.965 0.374 5.226 5.715 5.966 6.212 6.691 1.001 4400 beta.toad[10] 4.845 0.369 4.126 4.601 4.845 5.088 5.580 1.002 1700 beta.toad[11] 4.973 0.371 4.221 4.729 4.977 5.228 5.686 1.001 4400 beta.toad[12] 5.628 0.375 4.900 5.375 5.625 5.884 6.380 1.001 3900 beta.toad[13] 4.479 0.373 3.751 4.224 4.480 4.721 5.218 1.001 2600 beta.toad[14] 1.985 0.413 1.177 1.709 1.986 2.256 2.792 1.001 3700 beta.toad[15] 2.826 0.419 2.017 2.542 2.829 3.112 3.657 1.001 3500 beta.toad[16] 1.296 0.410 0.500 1.025 1.297 1.570 2.097 1.001 3300 beta.toad[17] 0.970 0.406 0.154 0.700 0.970 1.248 1.744 1.001 4400 beta.toad[18] 0.285 0.418 -0.532 0.004 0.289 0.560 1.084 1.001 4400 beta.toad[19] 2.189 0.413 1.375 1.909 2.194 2.465 2.993 1.001 4400 beta.toad[20] 1.590 0.418 0.772 1.310 1.587 1.870 2.406 1.001 4400 beta.toad[21] 1.259 0.411 0.451 0.992 1.251 1.539 2.061 1.001 3100 g0 4.937 0.357 4.213 4.699 4.940 5.181 5.625 1.001 3500 gamma.breath[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 gamma.breath[2] -3.396 0.589 -4.548 -3.790 -3.399 -3.007 -2.237 1.001 4400 sd.Breath 2.402 0.416 1.582 2.126 2.404 2.680 3.216 1.001 4400 sd.BreathO2level 1.483 0.206 1.080 1.344 1.486 1.621 1.889 1.002 2000 sd.O2level 0.844 0.096 0.662 0.780 0.845 0.908 1.031 1.002 2100 sd.Resid 1.483 0.000 1.483 1.483 1.483 1.483 1.483 1.000 1 sd.Toad 0.883 0.083 0.729 0.828 0.880 0.934 1.057 1.002 1400 sigma.breath 49.572 28.985 2.350 24.101 49.438 74.781 97.415 1.001 4400 sigma.breatho2level 0.979 0.517 0.366 0.663 0.875 1.166 2.224 1.001 4400 sigma.o2level 0.969 0.430 0.465 0.695 0.868 1.123 2.018 1.001 3500 sigma.res 0.881 0.056 0.779 0.842 0.878 0.916 1.000 1.001 4400 sigma.toad 0.944 0.188 0.650 0.811 0.920 1.050 1.369 1.001 4400 deviance 432.451 10.234 414.872 425.254 431.611 438.804 454.837 1.001 4400 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 52.4 and DIC = 484.8 DIC is an estimate of expected predictive error (lower deviance is better).Matrix parameterization JAGS codelibrary(R2jags) print(mullens.r2jags.b)
Inference for Bugs model at "../downloads/BUGSscripts/ws9.4bQ4.1b.txt", fit using jags, 3 chains, each with 1e+05 iterations (first 3000 discarded), n.thin = 100 n.sims = 2910 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff beta[1] 4.948 0.359 4.258 4.707 4.943 5.186 5.655 1.001 2900 beta[2] -3.405 0.571 -4.513 -3.784 -3.395 -3.035 -2.283 1.001 2900 beta[3] -0.219 0.346 -0.910 -0.447 -0.219 0.019 0.466 1.001 2900 beta[4] -0.549 0.349 -1.238 -0.783 -0.550 -0.315 0.143 1.001 2300 beta[5] -0.868 0.348 -1.580 -1.105 -0.866 -0.628 -0.192 1.001 2900 beta[6] -1.551 0.344 -2.226 -1.780 -1.549 -1.315 -0.886 1.001 2900 beta[7] -1.472 0.351 -2.161 -1.703 -1.472 -1.240 -0.784 1.001 2900 beta[8] -2.248 0.344 -2.920 -2.477 -2.248 -2.014 -1.585 1.001 2900 beta[9] -2.466 0.344 -3.144 -2.701 -2.473 -2.226 -1.801 1.001 2900 beta[10] 1.050 0.545 -0.044 0.698 1.046 1.404 2.137 1.001 2400 beta[11] 2.337 0.550 1.257 1.968 2.340 2.721 3.440 1.002 1400 beta[12] 2.383 0.568 1.256 2.018 2.377 2.764 3.502 1.001 2900 beta[13] 3.307 0.552 2.221 2.950 3.297 3.672 4.382 1.001 2900 beta[14] 2.994 0.564 1.886 2.631 2.995 3.368 4.078 1.001 2900 beta[15] 3.637 0.559 2.573 3.263 3.635 4.014 4.724 1.001 2900 beta[16] 3.474 0.559 2.391 3.091 3.472 3.847 4.566 1.001 2900 gamma[1] 0.428 0.426 -0.370 0.146 0.425 0.705 1.270 1.001 2500 gamma[2] -0.642 0.394 -1.442 -0.908 -0.641 -0.373 0.115 1.001 2900 gamma[3] 1.269 0.438 0.422 0.972 1.258 1.564 2.159 1.002 2600 gamma[4] 1.356 0.388 0.599 1.103 1.350 1.607 2.124 1.001 2900 gamma[5] -0.253 0.392 -1.047 -0.506 -0.252 0.009 0.510 1.001 2900 gamma[6] -0.551 0.392 -1.339 -0.814 -0.549 -0.282 0.196 1.001 2900 gamma[7] 1.419 0.399 0.640 1.149 1.418 1.685 2.202 1.001 2600 gamma[8] -0.261 0.431 -1.119 -0.552 -0.256 0.036 0.561 1.001 2400 gamma[9] -0.802 0.392 -1.554 -1.073 -0.802 -0.540 -0.031 1.001 2900 gamma[10] -0.569 0.428 -1.415 -0.854 -0.563 -0.288 0.299 1.001 2900 gamma[11] -1.253 0.391 -2.021 -1.510 -1.256 -0.984 -0.510 1.001 2200 gamma[12] -0.489 0.384 -1.253 -0.752 -0.482 -0.225 0.250 1.002 1400 gamma[13] 1.010 0.397 0.219 0.741 1.012 1.265 1.807 1.001 2000 gamma[14] -0.097 0.389 -0.868 -0.350 -0.096 0.165 0.650 1.000 2900 gamma[15] 0.029 0.379 -0.718 -0.224 0.028 0.289 0.772 1.001 2900 gamma[16] 0.677 0.381 -0.055 0.431 0.672 0.930 1.431 1.001 2900 gamma[17] -1.261 0.437 -2.112 -1.559 -1.253 -0.964 -0.409 1.001 2900 gamma[18] -0.464 0.386 -1.252 -0.713 -0.459 -0.205 0.286 1.001 2900 gamma[19] 0.642 0.430 -0.212 0.354 0.628 0.939 1.503 1.001 2900 gamma[20] 0.031 0.425 -0.817 -0.258 0.037 0.311 0.850 1.001 2900 gamma[21] -0.281 0.434 -1.139 -0.573 -0.282 -0.012 0.611 1.001 2900 sigma.res 0.878 0.055 0.778 0.839 0.876 0.911 0.994 1.001 2900 sigma.toad 0.942 0.181 0.651 0.816 0.918 1.044 1.364 1.003 980 deviance 430.909 9.736 414.255 424.034 430.244 437.014 452.334 1.001 2900 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 47.4 and DIC = 478.3 DIC is an estimate of expected predictive error (lower deviance is better).Matrix parameterization STAN codelibrary(R2jags) print(mullens.rstan.a)
Inference for Stan model: rstanString. 3 chains, each with iter=10000; warmup=3000; thin=10; post-warmup draws per chain=700, total post-warmup draws=2100. mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat beta[1] 4.94 0.01 0.36 4.23 4.70 4.94 5.18 5.62 1824 1 beta[2] -3.40 0.01 0.57 -4.50 -3.79 -3.40 -3.02 -2.27 1878 1 beta[3] -0.23 0.01 0.34 -0.89 -0.46 -0.23 -0.01 0.43 1814 1 beta[4] -0.56 0.01 0.35 -1.22 -0.79 -0.56 -0.32 0.11 1956 1 beta[5] -0.86 0.01 0.34 -1.52 -1.09 -0.87 -0.64 -0.18 1924 1 beta[6] -1.55 0.01 0.34 -2.23 -1.78 -1.55 -1.32 -0.88 2100 1 beta[7] -1.47 0.01 0.35 -2.13 -1.70 -1.46 -1.23 -0.79 2004 1 beta[8] -2.26 0.01 0.34 -2.92 -2.47 -2.25 -2.03 -1.57 2041 1 beta[9] -2.46 0.01 0.35 -3.11 -2.70 -2.46 -2.23 -1.78 1612 1 beta[10] 1.07 0.01 0.54 0.01 0.71 1.06 1.44 2.14 1856 1 beta[11] 2.35 0.01 0.55 1.28 1.97 2.35 2.74 3.46 1893 1 beta[12] 2.37 0.01 0.54 1.32 2.01 2.37 2.73 3.41 1989 1 beta[13] 3.31 0.01 0.54 2.27 2.93 3.31 3.69 4.39 2100 1 beta[14] 2.98 0.01 0.53 1.96 2.59 2.99 3.34 4.05 2100 1 beta[15] 3.64 0.01 0.55 2.54 3.25 3.63 4.01 4.70 2100 1 beta[16] 3.46 0.01 0.55 2.42 3.10 3.47 3.83 4.53 2100 1 gamma[1] 0.45 0.01 0.44 -0.39 0.16 0.45 0.74 1.29 1795 1 gamma[2] -0.63 0.01 0.40 -1.46 -0.89 -0.64 -0.38 0.17 1540 1 gamma[3] 1.28 0.01 0.45 0.43 0.99 1.28 1.57 2.16 1806 1 gamma[4] 1.37 0.01 0.39 0.63 1.11 1.36 1.63 2.16 2094 1 gamma[5] -0.22 0.01 0.39 -0.96 -0.49 -0.23 0.04 0.53 2003 1 gamma[6] -0.54 0.01 0.39 -1.30 -0.80 -0.55 -0.28 0.23 1843 1 gamma[7] 1.43 0.01 0.38 0.67 1.17 1.43 1.68 2.16 2075 1 gamma[8] -0.25 0.01 0.44 -1.10 -0.54 -0.24 0.04 0.62 2100 1 gamma[9] -0.78 0.01 0.40 -1.58 -1.04 -0.77 -0.50 -0.02 1918 1 gamma[10] -0.57 0.01 0.43 -1.44 -0.85 -0.57 -0.28 0.24 1888 1 gamma[11] -1.23 0.01 0.38 -1.99 -1.48 -1.23 -0.97 -0.48 1878 1 gamma[12] -0.48 0.01 0.38 -1.20 -0.74 -0.48 -0.21 0.26 1819 1 gamma[13] 1.02 0.01 0.40 0.27 0.76 1.02 1.29 1.81 1986 1 gamma[14] -0.08 0.01 0.39 -0.84 -0.34 -0.07 0.17 0.64 1883 1 gamma[15] 0.03 0.01 0.38 -0.72 -0.23 0.03 0.29 0.82 1870 1 gamma[16] 0.68 0.01 0.39 -0.10 0.42 0.67 0.94 1.43 1861 1 gamma[17] -1.25 0.01 0.45 -2.15 -1.54 -1.24 -0.95 -0.38 1987 1 gamma[18] -0.45 0.01 0.39 -1.23 -0.69 -0.45 -0.19 0.29 1694 1 gamma[19] 0.65 0.01 0.45 -0.19 0.36 0.65 0.95 1.55 2011 1 gamma[20] 0.06 0.01 0.45 -0.81 -0.23 0.06 0.34 0.95 2042 1 gamma[21] -0.28 0.01 0.43 -1.11 -0.56 -0.28 0.00 0.58 1975 1 sigma 0.88 0.00 0.05 0.78 0.84 0.88 0.91 1.00 2100 1 sigma_Z 0.94 0.00 0.19 0.65 0.81 0.91 1.05 1.40 2100 1 lp__ -69.80 0.11 4.97 -80.69 -72.81 -69.40 -66.24 -61.43 2080 1 Samples were drawn using NUTS(diag_e) at Sat Jan 31 08:07:18 2015. For each parameter, n_eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence, Rhat=1). - Figure time
Matrix parameterization JAGS code
nms <- colnames(mullens.mcmc.b) pred <- grep('beta',nms) coefs <- mullens.mcmc.b[,pred] newdata <- expand.grid(BREATH=levels(mullens$BREATH), O2LEVEL=levels(mullens$O2LEVEL)) Xmat <- model.matrix(~BREATH*O2LEVEL, newdata) pred <- coefs %*% t(Xmat) library(plyr) newdata <- cbind(newdata, adply(pred, 2, function(x) { data.frame(Median=median(x), HPDinterval(as.mcmc(x)), HPDinterval(as.mcmc(x), p=0.68)) })) newdata
BREATH O2LEVEL X1 Median lower upper lower.1 upper.1 1 buccal 0 1 4.943 4.2389 5.618 4.561 5.269 2 lung 0 2 1.550 0.6998 2.466 1.132 2.001 3 buccal 5 3 4.728 4.0285 5.432 4.371 5.082 4 lung 5 4 2.375 1.5123 3.301 1.848 2.747 5 buccal 10 5 4.395 3.7066 5.086 4.007 4.726 6 lung 10 6 3.325 2.4879 4.218 2.871 3.765 7 buccal 15 7 4.085 3.3682 4.791 3.682 4.406 8 lung 15 8 3.059 2.1765 3.968 2.571 3.469 9 buccal 20 9 3.389 2.6788 4.076 3.011 3.718 10 lung 20 10 3.301 2.3525 4.161 2.842 3.722 11 buccal 30 11 3.480 2.7380 4.156 3.168 3.891 12 lung 30 12 3.066 2.1262 3.906 2.615 3.502 13 buccal 40 13 2.698 2.0185 3.435 2.332 3.018 14 lung 40 14 2.923 2.0682 3.855 2.511 3.384 15 buccal 50 15 2.470 1.7995 3.203 2.113 2.827 16 lung 50 16 2.548 1.7116 3.480 2.104 3.005
library(ggplot2) p1 <-ggplot(newdata, aes(y=Median, x=as.integer(O2LEVEL), fill=BREATH)) + geom_line(aes(linetype=BREATH)) + #geom_line()+ geom_linerange(aes(ymin=lower, ymax=upper), show_guide=FALSE)+ geom_linerange(aes(ymin=lower.1, ymax=upper.1), size=2,show_guide=FALSE)+ geom_point(size=3, shape=21)+ scale_y_continuous(expression(Frequency~of~buccal~breathing))+ scale_x_continuous('Oxygen concentration')+ scale_fill_manual('Breath type', breaks=c('buccal','lung'), values=c('black','white'), labels=c('Buccal','Lung'))+ scale_linetype_manual('Breath type', breaks=c('buccal','lung'), values=c('solid','dashed'), labels=c('Buccal','Lung'))+ theme_classic() + theme(legend.justification=c(1,0), legend.position=c(1,0), legend.key.width=unit(3,'lines'), axis.title.y=element_text(vjust=2, size=rel(1.2)), axis.title.x=element_text(vjust=-1, size=rel(1.2)), plot.margin=unit(c(0.5,0.5,1,2),'lines')) mullens.sd$Source <- factor(mullens.sd$Source, levels=c("Resid","BREATH:O2LEVEL","O2LEVEL","TOAD","BREATH"), labels=c("Residuals", "Breath:O2", "O2","Toad","Breath")) mullens.sd$Perc <- 100*mullens.sd$Median/sum(mullens.sd$Median) p2<-ggplot(mullens.sd,aes(y=Source, x=Median))+ geom_vline(xintercept=0,linetype="dashed")+ geom_hline(xintercept=0)+ scale_x_continuous("Finite population \nvariance components (sd)")+ geom_errorbarh(aes(xmin=lower.1, xmax=upper.1), height=0, size=1)+ geom_errorbarh(aes(xmin=lower, xmax=upper), height=0, size=1.5)+ geom_point(size=3)+ geom_text(aes(label=sprintf("(%4.1f%%)",Perc),vjust=-1))+ theme_classic()+ theme(axis.line=element_blank(),axis.title.y=element_blank(), axis.text.y=element_text(size=12,hjust=1)) library(gridExtra) grid.arrange(p1, p2, ncol=2)
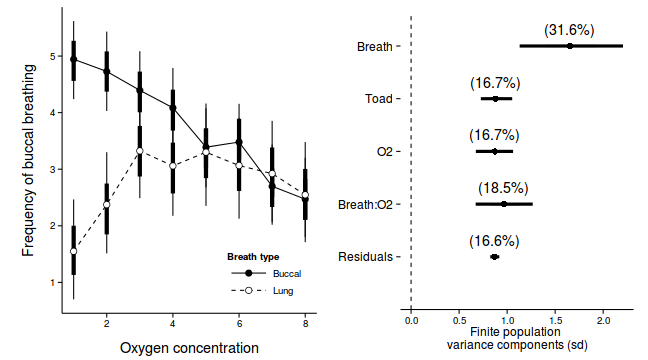
- As an alternative, we could model oxygen concentration as a numeric...
Matrix parameterization JAGS codeWe can perform generate the finite-population standard deviation posteriors within R
modelString=" model { #Likelihood for (i in 1:n) { y[i]~dnorm(mu[i],tau.res) mu[i] <- inprod(beta[],X[i,]) + inprod(gamma[],Z[i,]) y.err[i] <- y[i] - mu[1] } #Priors and derivatives for (i in 1:nZ) { gamma[i] ~ dnorm(0,tau.toad) } for (i in 1:nX) { beta[i] ~ dnorm(0,1.0E-06) } tau.res <- pow(sigma.res,-2) sigma.res <- z/sqrt(chSq) z ~ dnorm(0, .0016)I(0,) chSq ~ dgamma(0.5, 0.5) tau.toad <- pow(sigma.toad,-2) sigma.toad <- z.toad/sqrt(chSq.toad) z.toad ~ dnorm(0, .0016)I(0,) chSq.toad ~ dgamma(0.5, 0.5) } " ## write the model to a text file (I suggest you alter the path to somewhere more relevant ## to your system!) writeLines(modelString,con="../downloads/BUGSscripts/ws9.4bQ4.6a.txt") Xmat <- model.matrix(~BREATH*iO2LEVEL, data=mullens) Zmat <- model.matrix(~-1+TOAD, data=mullens) mullens.list <- list(y=mullens$SFREQBUC, X=Xmat, nX=ncol(Xmat), Z=Zmat, nZ=ncol(Zmat), n=nrow(mullens) ) params <- c("beta","gamma", "sigma.res","sigma.toad") burnInSteps = 3000 nChains = 3 numSavedSteps = 3000 thinSteps = 100 nIter = ceiling((numSavedSteps * thinSteps)/nChains) library(R2jags) library(coda) ## mullens.r2jags.d <- jags(data=mullens.list, inits=NULL, # since there are three chains parameters.to.save=params, model.file="../downloads/BUGSscripts/ws9.4bQ4.6a.txt", n.chains=3, n.iter=nIter, n.burnin=burnInSteps, n.thin=thinSteps )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 5045 Initializing model
print(mullens.r2jags.d)
Inference for Bugs model at "../downloads/BUGSscripts/ws9.4bQ4.6a.txt", fit using jags, 3 chains, each with 1e+05 iterations (first 3000 discarded), n.thin = 100 n.sims = 2910 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff beta[1] 4.848 0.308 4.234 4.641 4.850 5.059 5.445 1.001 2900 beta[2] -2.296 0.503 -3.254 -2.639 -2.288 -1.982 -1.287 1.001 2900 beta[3] -0.051 0.006 -0.062 -0.054 -0.051 -0.047 -0.040 1.002 1900 beta[4] 0.062 0.009 0.043 0.055 0.062 0.067 0.080 1.003 1400 gamma[1] 0.417 0.445 -0.482 0.115 0.424 0.719 1.277 1.001 2900 gamma[2] -0.636 0.397 -1.417 -0.895 -0.642 -0.369 0.130 1.001 2900 gamma[3] 1.252 0.449 0.365 0.957 1.244 1.548 2.158 1.001 2900 gamma[4] 1.342 0.402 0.573 1.075 1.336 1.602 2.170 1.001 2900 gamma[5] -0.233 0.403 -1.034 -0.494 -0.229 0.038 0.558 1.001 2900 gamma[6] -0.538 0.398 -1.297 -0.803 -0.543 -0.278 0.224 1.001 2900 gamma[7] 1.418 0.410 0.624 1.131 1.416 1.684 2.258 1.001 2900 gamma[8] -0.272 0.439 -1.148 -0.568 -0.267 0.021 0.587 1.002 1100 gamma[9] -0.778 0.398 -1.567 -1.040 -0.769 -0.513 0.000 1.001 2900 gamma[10] -0.584 0.445 -1.473 -0.880 -0.585 -0.277 0.264 1.001 2200 gamma[11] -1.226 0.403 -2.025 -1.504 -1.219 -0.949 -0.449 1.001 2900 gamma[12] -0.487 0.392 -1.236 -0.758 -0.487 -0.221 0.270 1.001 2900 gamma[13] 1.012 0.399 0.253 0.747 1.000 1.270 1.802 1.001 2900 gamma[14] -0.085 0.399 -0.875 -0.342 -0.083 0.185 0.717 1.001 2900 gamma[15] 0.031 0.396 -0.744 -0.232 0.030 0.289 0.810 1.001 2900 gamma[16] 0.673 0.403 -0.067 0.401 0.664 0.931 1.490 1.001 2900 gamma[17] -1.263 0.456 -2.202 -1.568 -1.257 -0.952 -0.383 1.001 2900 gamma[18] -0.457 0.404 -1.275 -0.728 -0.457 -0.186 0.322 1.001 2900 gamma[19] 0.624 0.443 -0.238 0.333 0.627 0.919 1.503 1.001 2900 gamma[20] 0.030 0.442 -0.849 -0.257 0.030 0.329 0.911 1.001 2900 gamma[21] -0.295 0.448 -1.187 -0.593 -0.298 0.004 0.583 1.001 2900 sigma.res 0.925 0.054 0.824 0.888 0.922 0.961 1.038 1.002 1000 sigma.toad 0.938 0.188 0.638 0.804 0.919 1.046 1.366 1.001 2900 deviance 449.164 7.815 436.212 443.536 448.455 454.081 466.831 1.001 2900 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 30.6 and DIC = 479.7 DIC is an estimate of expected predictive error (lower deviance is better).mullens.mcmc.d <- as.mcmc(mullens.r2jags.d$BUGSoutput$sims.matrix)
View full codenms <- colnames(mullens.mcmc.d) pred <- grep('beta|gamma',nms) coefs <- mullens.mcmc.d[,pred] newdata <- mullens Amat <- cbind(model.matrix(~BREATH*iO2LEVEL, newdata),model.matrix(~-1+TOAD, newdata)) pred <- coefs %*% t(Amat) y.err <- t(mullens$SFREQBUC - t(pred)) a<-apply(y.err, 1,sd) resid.sd <- data.frame(Source='Resid',Median=median(a), HPDinterval(as.mcmc(a)), HPDinterval(as.mcmc(a), p=0.68)) ## Fixed effects nms <- colnames(mullens.mcmc.d) mullens.mcmc.d.beta <- mullens.mcmc.d[,grep('beta',nms)] head(mullens.mcmc.d.beta)
Markov Chain Monte Carlo (MCMC) output: Start = 1 End = 7 Thinning interval = 1 beta[1] beta[2] beta[3] beta[4] [1,] 4.674 -2.208 -0.05177 0.06361 [2,] 5.172 -2.802 -0.06305 0.06144 [3,] 5.563 -2.930 -0.06357 0.06296 [4,] 4.651 -2.148 -0.04643 0.05753 [5,] 5.183 -2.749 -0.05130 0.06486 [6,] 4.836 -1.440 -0.05132 0.06433 [7,] 4.938 -2.385 -0.04854 0.06522assign <- attr(Xmat, 'assign') assign
[1] 0 1 2 3
nms <- attr(terms(model.frame(~BREATH*iO2LEVEL, mullens)),'term.labels') nms
[1] "BREATH" "iO2LEVEL" "BREATH:iO2LEVEL"
fixed.sd <- NULL for (i in 1:max(assign)) { w <- which(i==assign) if (length(w)==1) fixed.sd<-cbind(fixed.sd, abs(mullens.mcmc.d.beta[,w])*sd(Xmat[,w])) else fixed.sd<-cbind(fixed.sd, apply(mullens.mcmc.d.beta[,w],1,sd)) } library(plyr) fixed.sd <-adply(fixed.sd,2,function(x) { data.frame(Median=median(x), HPDinterval(as.mcmc(x)), HPDinterval(as.mcmc(x), p=0.68)) }) fixed.sd<- cbind(Source=nms, fixed.sd) # Variances nms <- colnames(mullens.mcmc.d) mullens.mcmc.d.gamma <- mullens.mcmc.d[,grep('gamma',nms)] assign <- attr(Zmat, 'assign') nms <- attr(terms(model.frame(~-1+TOAD, mullens)),'term.labels') random.sd <- NULL for (i in 1:max(assign)) { w <- which(i==assign) random.sd<-cbind(random.sd, apply(mullens.mcmc.d.gamma[,w],1,sd)) } library(plyr) random.sd <-adply(random.sd,2,function(x) { data.frame(Median=median(x), HPDinterval(as.mcmc(x)), HPDinterval(as.mcmc(x), p=0.68)) }) random.sd<- cbind(Source=nms, random.sd) (mullens.sd <- rbind(subset(fixed.sd, select=-X1), subset(random.sd, select=-X1), resid.sd))
Source Median lower upper lower.1 upper.1 1 BREATH 1.1145 0.6544 1.6077 0.9031 1.3725 2 iO2LEVEL 0.8339 0.6455 1.0092 0.7504 0.9291 3 BREATH:iO2LEVEL 0.8914 0.6501 1.1791 0.7742 1.0314 4 TOAD 0.8740 0.6970 1.0544 0.7874 0.9527 var1 Resid 0.9164 0.8814 0.9603 0.8941 0.9336
- Better still, what about a polynomial (2nd order: rather than simple linear).
Matrix parameterization JAGS codeWe can perform generate the finite-population standard deviation posteriors within R
modelString=" model { #Likelihood for (i in 1:n) { y[i]~dnorm(mu[i],tau.res) mu[i] <- inprod(beta[],X[i,]) + inprod(gamma[],Z[i,]) y.err[i] <- y[i] - mu[1] } #Priors and derivatives for (i in 1:nZ) { gamma[i] ~ dnorm(0,tau.toad) } for (i in 1:nX) { beta[i] ~ dnorm(0,1.0E-06) } tau.res <- pow(sigma.res,-2) sigma.res <- z/sqrt(chSq) z ~ dnorm(0, .0016)I(0,) chSq ~ dgamma(0.5, 0.5) tau.toad <- pow(sigma.toad,-2) sigma.toad <- z.toad/sqrt(chSq.toad) z.toad ~ dnorm(0, .0016)I(0,) chSq.toad ~ dgamma(0.5, 0.5) } " ## write the model to a text file (I suggest you alter the path to somewhere more relevant ## to your system!) writeLines(modelString,con="../downloads/BUGSscripts/ws9.4bQ4.7a.txt") Xmat <- model.matrix(~BREATH*poly(iO2LEVEL,2), data=mullens) Zmat <- model.matrix(~-1+TOAD, data=mullens) mullens.list <- list(y=mullens$SFREQBUC, X=Xmat, nX=ncol(Xmat), Z=Zmat, nZ=ncol(Zmat), n=nrow(mullens) ) params <- c("beta","gamma", "sigma.res","sigma.toad") burnInSteps = 3000 nChains = 3 numSavedSteps = 3000 thinSteps = 100 nIter = ceiling((numSavedSteps * thinSteps)/nChains) library(R2jags) library(coda) ## mullens.r2jags.f <- jags(data=mullens.list, inits=NULL, # since there are three chains parameters.to.save=params, model.file="../downloads/BUGSscripts/ws9.4bQ4.7a.txt", n.chains=3, n.iter=nIter, n.burnin=burnInSteps, n.thin=thinSteps )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 5383 Initializing model
print(mullens.r2jags.f)
Inference for Bugs model at "../downloads/BUGSscripts/ws9.4bQ4.7a.txt", fit using jags, 3 chains, each with 1e+05 iterations (first 3000 discarded), n.thin = 100 n.sims = 2910 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff beta[1] 3.779 0.280 3.222 3.599 3.776 3.961 4.343 1.001 2900 beta[2] -1.010 0.453 -1.892 -1.294 -1.022 -0.722 -0.119 1.001 2900 beta[3] -10.777 1.122 -12.936 -11.514 -10.766 -10.037 -8.550 1.002 1100 beta[4] 1.296 1.115 -0.934 0.551 1.318 2.056 3.475 1.001 2900 beta[5] 13.044 1.813 9.489 11.802 13.101 14.257 16.499 1.001 2900 beta[6] -7.228 1.792 -10.737 -8.416 -7.236 -6.051 -3.586 1.001 2900 gamma[1] 0.443 0.429 -0.390 0.157 0.453 0.731 1.295 1.001 2900 gamma[2] -0.636 0.392 -1.433 -0.896 -0.635 -0.374 0.133 1.001 2900 gamma[3] 1.282 0.431 0.453 0.981 1.276 1.565 2.138 1.001 2900 gamma[4] 1.353 0.383 0.599 1.094 1.356 1.609 2.095 1.001 2900 gamma[5] -0.252 0.388 -1.005 -0.505 -0.246 0.001 0.521 1.001 2900 gamma[6] -0.556 0.386 -1.307 -0.807 -0.566 -0.298 0.221 1.001 2900 gamma[7] 1.418 0.384 0.675 1.168 1.409 1.664 2.198 1.001 2900 gamma[8] -0.255 0.440 -1.130 -0.539 -0.250 0.036 0.585 1.001 2900 gamma[9] -0.804 0.393 -1.560 -1.062 -0.793 -0.538 -0.057 1.001 2900 gamma[10] -0.586 0.438 -1.440 -0.864 -0.589 -0.294 0.269 1.002 1400 gamma[11] -1.242 0.397 -2.038 -1.510 -1.239 -0.972 -0.479 1.001 2900 gamma[12] -0.499 0.386 -1.281 -0.761 -0.492 -0.248 0.254 1.001 2900 gamma[13] 1.012 0.384 0.284 0.753 1.006 1.263 1.793 1.001 2900 gamma[14] -0.102 0.397 -0.895 -0.362 -0.100 0.160 0.688 1.001 2900 gamma[15] 0.013 0.383 -0.731 -0.232 0.008 0.268 0.780 1.002 1600 gamma[16] 0.678 0.385 -0.101 0.421 0.676 0.928 1.441 1.001 2900 gamma[17] -1.270 0.434 -2.146 -1.550 -1.261 -0.983 -0.422 1.001 2600 gamma[18] -0.466 0.389 -1.253 -0.717 -0.459 -0.211 0.294 1.001 2900 gamma[19] 0.643 0.433 -0.196 0.347 0.640 0.934 1.496 1.001 2900 gamma[20] 0.046 0.430 -0.812 -0.247 0.046 0.342 0.879 1.001 2500 gamma[21] -0.281 0.438 -1.139 -0.569 -0.281 0.022 0.541 1.001 2900 sigma.res 0.873 0.052 0.782 0.837 0.870 0.906 0.984 1.001 2900 sigma.toad 0.939 0.184 0.652 0.809 0.914 1.043 1.366 1.001 2900 deviance 430.191 7.965 416.104 424.602 429.731 435.046 447.182 1.001 2000 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 31.7 and DIC = 461.9 DIC is an estimate of expected predictive error (lower deviance is better).mullens.mcmc.f <- as.mcmc(mullens.r2jags.f$BUGSoutput$sims.matrix)
View full codenms <- colnames(mullens.mcmc.f) pred <- grep('beta|gamma',nms) coefs <- mullens.mcmc.f[,pred] newdata <- mullens Amat <- cbind(model.matrix(~BREATH*poly(iO2LEVEL,2), newdata),model.matrix(~-1+TOAD, newdata)) pred <- coefs %*% t(Amat) y.err <- t(mullens$SFREQBUC - t(pred)) a<-apply(y.err, 1,sd) resid.sd <- data.frame(Source='Resid',Median=median(a), HPDinterval(as.mcmc(a)), HPDinterval(as.mcmc(a), p=0.68)) ## Fixed effects nms <- colnames(mullens.mcmc.f) mullens.mcmc.f.beta <- mullens.mcmc.f[,grep('beta',nms)] head(mullens.mcmc.f.beta)
Markov Chain Monte Carlo (MCMC) output: Start = 1 End = 7 Thinning interval = 1 beta[1] beta[2] beta[3] beta[4] beta[5] beta[6] [1,] 3.543 -0.5386 -9.886 0.9750 12.133 -5.675 [2,] 3.868 -0.6460 -10.644 0.2724 13.265 -4.126 [3,] 3.840 -0.6493 -8.714 1.5150 9.811 -10.104 [4,] 3.772 -1.0448 -10.627 2.7407 12.849 -8.501 [5,] 3.674 -1.3568 -10.291 1.2977 9.863 -9.403 [6,] 3.880 -0.8706 -10.255 0.4072 12.889 -4.371 [7,] 4.048 -1.4604 -11.777 1.0588 13.864 -9.134assign <- attr(Xmat, 'assign') assign
[1] 0 1 2 2 3 3
nms <- attr(terms(model.frame(~BREATH*poly(iO2LEVEL,2), mullens)),'term.labels') nms
[1] "BREATH" "poly(iO2LEVEL, 2)" "BREATH:poly(iO2LEVEL, 2)"
fixed.sd <- NULL for (i in 1:max(assign)) { w <- which(i==assign) if (length(w)==1) fixed.sd<-cbind(fixed.sd, abs(mullens.mcmc.f.beta[,w])*sd(Xmat[,w])) else fixed.sd<-cbind(fixed.sd, apply(mullens.mcmc.f.beta[,w],1,sd)) } library(plyr) fixed.sd <-adply(fixed.sd,2,function(x) { data.frame(Median=median(x), HPDinterval(as.mcmc(x)), HPDinterval(as.mcmc(x), p=0.68)) }) fixed.sd<- cbind(Source=nms, fixed.sd) # Variances nms <- colnames(mullens.mcmc.f) mullens.mcmc.f.gamma <- mullens.mcmc.f[,grep('gamma',nms)] assign <- attr(Zmat, 'assign') nms <- attr(terms(model.frame(~-1+TOAD, mullens)),'term.labels') random.sd <- NULL for (i in 1:max(assign)) { w <- which(i==assign) random.sd<-cbind(random.sd, apply(mullens.mcmc.f.gamma[,w],1,sd)) } library(plyr) random.sd <-adply(random.sd,2,function(x) { data.frame(Median=median(x), HPDinterval(as.mcmc(x)), HPDinterval(as.mcmc(x), p=0.68)) }) random.sd<- cbind(Source=nms, random.sd) (mullens.sd <- rbind(subset(fixed.sd, select=-X1), subset(random.sd, select=-X1), resid.sd))
Source Median lower upper lower.1 upper.1 1 BREATH 0.4977 0.05243 0.8759 0.2554 0.6765 2 poly(iO2LEVEL, 2) 8.5430 6.33552 10.7267 7.5627 9.8334 3 BREATH:poly(iO2LEVEL, 2) 14.3771 10.92213 17.9611 12.7608 16.3635 4 TOAD 0.8794 0.72207 1.0400 0.7985 0.9510 var1 Resid 0.8663 0.82980 0.9053 0.8450 0.8828
- Matrix parameterization - random intercepts model (STAN)
Matrix parameterization STAN coderstanString=" data{ int n; int nZ; int nX; vector [n] y; matrix [n,nX] X; matrix [n,nZ] Z; vector [nX] a0; matrix [nX,nX] A0; } parameters{ vector [nX] beta; real<lower=0> sigma; vector [nZ] gamma; real<lower=0> sigma_Z; } transformed parameters { vector [n] mu; mu <- X*beta + Z*gamma; } model{ // Priors beta ~ multi_normal(a0,A0); gamma ~ normal( 0 , sigma_Z ); sigma_Z ~ cauchy(0,25); sigma ~ cauchy(0,25); y ~ normal( mu , sigma ); } generated quantities { vector [n] y_err; real<lower=0> sd_Resid; y_err <- y - mu; sd_Resid <- sd(y_err); } " #sort the data set so that the copper treatments are in a more logical order Xmat <- model.matrix(~BREATH*poly(iO2LEVEL,2), data=mullens) Zmat <- model.matrix(~-1+TOAD, data=mullens) mullens.list <- list(y=mullens$SFREQBUC, X=Xmat, nX=ncol(Xmat), Z=Zmat, nZ=ncol(Zmat), n=nrow(mullens), a0=rep(0,ncol(Xmat)), A0=diag(100000,ncol(Xmat)) ) params <- c("beta","gamma", "sigma","sigma_Z") burnInSteps = 3000 nChains = 3 numSavedSteps = 3000 thinSteps = 10 nIter = ceiling((numSavedSteps * thinSteps)/nChains) library(rstan) library(coda) ## mullens.rstan.g <- stan(data=mullens.list, model_code=rstanString, pars=c("beta","gamma","sigma","sigma_Z"), chains=nChains, iter=nIter, warmup=burnInSteps, thin=thinSteps, save_dso=TRUE )
TRANSLATING MODEL 'rstanString' FROM Stan CODE TO C++ CODE NOW. COMPILING THE C++ CODE FOR MODEL 'rstanString' NOW. SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 1). Iteration: 1 / 10000 [ 0%] (Warmup) Iteration: 1000 / 10000 [ 10%] (Warmup) Iteration: 2000 / 10000 [ 20%] (Warmup) Iteration: 3000 / 10000 [ 30%] (Warmup) Iteration: 3001 / 10000 [ 30%] (Sampling) Iteration: 4000 / 10000 [ 40%] (Sampling) Iteration: 5000 / 10000 [ 50%] (Sampling) Iteration: 6000 / 10000 [ 60%] (Sampling) Iteration: 7000 / 10000 [ 70%] (Sampling) Iteration: 8000 / 10000 [ 80%] (Sampling) Iteration: 9000 / 10000 [ 90%] (Sampling) Iteration: 10000 / 10000 [100%] (Sampling) # Elapsed Time: 3.58 seconds (Warm-up) # 8.76 seconds (Sampling) # 12.34 seconds (Total) SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 2). Iteration: 1 / 10000 [ 0%] (Warmup) Iteration: 1000 / 10000 [ 10%] (Warmup) Iteration: 2000 / 10000 [ 20%] (Warmup) Iteration: 3000 / 10000 [ 30%] (Warmup) Iteration: 3001 / 10000 [ 30%] (Sampling) Iteration: 4000 / 10000 [ 40%] (Sampling) Iteration: 5000 / 10000 [ 50%] (Sampling) Iteration: 6000 / 10000 [ 60%] (Sampling) Iteration: 7000 / 10000 [ 70%] (Sampling) Iteration: 8000 / 10000 [ 80%] (Sampling) Iteration: 9000 / 10000 [ 90%] (Sampling) Iteration: 10000 / 10000 [100%] (Sampling) # Elapsed Time: 3.73 seconds (Warm-up) # 7.78 seconds (Sampling) # 11.51 seconds (Total) SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 3). Iteration: 1 / 10000 [ 0%] (Warmup) Iteration: 1000 / 10000 [ 10%] (Warmup) Iteration: 2000 / 10000 [ 20%] (Warmup) Iteration: 3000 / 10000 [ 30%] (Warmup) Iteration: 3001 / 10000 [ 30%] (Sampling) Iteration: 4000 / 10000 [ 40%] (Sampling) Iteration: 5000 / 10000 [ 50%] (Sampling) Iteration: 6000 / 10000 [ 60%] (Sampling) Iteration: 7000 / 10000 [ 70%] (Sampling) Iteration: 8000 / 10000 [ 80%] (Sampling) Iteration: 9000 / 10000 [ 90%] (Sampling) Iteration: 10000 / 10000 [100%] (Sampling) # Elapsed Time: 3.19 seconds (Warm-up) # 7.28 seconds (Sampling) # 10.47 seconds (Total)
print(mullens.rstan.g)
Inference for Stan model: rstanString. 3 chains, each with iter=10000; warmup=3000; thin=10; post-warmup draws per chain=700, total post-warmup draws=2100. mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat beta[1] 3.76 0.01 0.28 3.21 3.58 3.76 3.94 4.34 1759 1 beta[2] -1.00 0.01 0.45 -1.90 -1.28 -1.00 -0.71 -0.12 1700 1 beta[3] -10.76 0.03 1.15 -13.07 -11.52 -10.74 -9.98 -8.57 1914 1 beta[4] 1.32 0.02 1.13 -0.89 0.56 1.31 2.06 3.57 2062 1 beta[5] 13.06 0.04 1.83 9.54 11.82 13.04 14.27 16.82 2100 1 beta[6] -7.21 0.04 1.76 -10.61 -8.42 -7.26 -6.02 -3.81 1973 1 gamma[1] 0.44 0.01 0.44 -0.42 0.15 0.43 0.72 1.35 1868 1 gamma[2] -0.62 0.01 0.37 -1.36 -0.87 -0.61 -0.38 0.08 1847 1 gamma[3] 1.28 0.01 0.45 0.41 0.98 1.28 1.56 2.18 1822 1 gamma[4] 1.37 0.01 0.39 0.61 1.10 1.38 1.64 2.12 1955 1 gamma[5] -0.23 0.01 0.38 -0.98 -0.49 -0.24 0.03 0.51 1933 1 gamma[6] -0.53 0.01 0.39 -1.33 -0.78 -0.52 -0.28 0.21 1881 1 gamma[7] 1.44 0.01 0.39 0.65 1.18 1.45 1.70 2.19 1901 1 gamma[8] -0.25 0.01 0.45 -1.13 -0.54 -0.25 0.02 0.65 1991 1 gamma[9] -0.78 0.01 0.37 -1.52 -1.03 -0.79 -0.53 -0.05 1910 1 gamma[10] -0.57 0.01 0.44 -1.43 -0.85 -0.59 -0.29 0.31 1933 1 gamma[11] -1.22 0.01 0.37 -1.96 -1.47 -1.20 -0.97 -0.51 1959 1 gamma[12] -0.49 0.01 0.38 -1.23 -0.75 -0.48 -0.23 0.25 1727 1 gamma[13] 1.03 0.01 0.38 0.28 0.78 1.02 1.28 1.79 1973 1 gamma[14] -0.09 0.01 0.39 -0.87 -0.34 -0.08 0.18 0.68 1899 1 gamma[15] 0.02 0.01 0.38 -0.71 -0.24 0.03 0.27 0.79 1921 1 gamma[16] 0.69 0.01 0.39 -0.11 0.45 0.70 0.95 1.43 1977 1 gamma[17] -1.27 0.01 0.45 -2.16 -1.56 -1.26 -0.98 -0.39 2013 1 gamma[18] -0.45 0.01 0.39 -1.24 -0.71 -0.45 -0.19 0.30 1801 1 gamma[19] 0.63 0.01 0.45 -0.24 0.33 0.63 0.93 1.56 1925 1 gamma[20] 0.05 0.01 0.44 -0.82 -0.23 0.04 0.34 0.88 1952 1 gamma[21] -0.28 0.01 0.44 -1.12 -0.57 -0.29 0.00 0.59 1950 1 sigma 0.87 0.00 0.05 0.78 0.84 0.87 0.91 0.98 1981 1 sigma_Z 0.93 0.00 0.18 0.64 0.81 0.91 1.04 1.36 2100 1 lp__ -69.37 0.09 4.18 -78.42 -72.00 -69.14 -66.51 -61.89 2001 1 Samples were drawn using NUTS(diag_e) at Sat Jan 31 14:46:33 2015. For each parameter, n_eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence, Rhat=1).mullens.mcmc.g <- rstan:::as.mcmc.list.stanfit(mullens.rstan.g) mullens.mcmc.df.g <- as.data.frame(extract(mullens.rstan.g))
