Tutorial 9.7b - Nested ANOVA (Bayesian)
14 Jan 2014
If you are completely ontop of the conceptual issues pertaining to Nested ANOVA, and just need to use this tutorial in order to learn about Nested ANOVA in R, you are invited to skip down to the section on Nested ANOVA in R.
Overview
You are strongly advised to review the information on the nested design in tutorial 9.7a. I am not going to duplicate the overview here.
Linear models
So called 'random effects' are modelled differently from 'fixed effects' in that rather than estimate their individual effects, we instead estimate the variability due to these 'random effects'. Since technically all variables in a Bayesian framework are random, some prefer to use the terms 'fixed effects' and 'varying effects'. As random factors typically represent 'random' selections of levels (such as a set of randomly selected sites), incorporated in order to account for the dependency structure (observations within sites are more likely to be correlated to one another - not strickly independent) to the data, we are not overly interested in the individual differences between levels of the 'varying' (random) factor. Instead (in addition to imposing a separate correlation structure within each nest), we want to know how much variability is attributed to this level of the design.
The linear models for two and three factor nested design are:
$$
\begin{align}
y_{ijk}&=\mu+\alpha_i + \beta_{j(i)} + \varepsilon_{ijk} &\hspace{2em} \varepsilon_{ijk}&\sim\mathcal{N}(0,\sigma^2), \hspace{1em}\beta_{j(i)}\sim\mathcal{N}(0,\sigma_B^2)\\
y_{ijkl}&=\mu+\alpha_i + \beta_{j(i)} + \gamma_{k(j(i))} + \varepsilon_{ijkl} &\hspace{2em} \varepsilon_{ijkl}&\sim\mathcal{N}(0,\sigma^2), \hspace{1em}\beta_{j(i)}\sim\mathcal{N}(0,\sigma_B^2), \hspace{1em}\gamma_{k(j(i))}\sim\mathcal{N}(0,\sigma_C^2)\\
\end{align}
$$
where $\mu$ is the overall mean, $\alpha$ is the effect of Factor A, $\beta$ is the variability of Factor B (nested within Factor A), $\gamma$ is the variability of Factor C (nested within Factor B) and $\varepsilon$ is the random unexplained or residual component that is assumed to be normally distributed with a mean of zero and a constant amount of standard deviation ($\sigma^2$).
The subscripts are iterators. For example, the $i$ represents the number of effects to be estimated for Factor A. Thus the first formula can be read as 'the $k$th observation of $y$ is drawn from a normal distribution (with a specific level of variability) and mean proposed to be determined by a base mean ($\mu$ - mean of the first treatment across all nests) plus the effect of the $i$th treatment effect plus the variability 'the model proposes that given a base mean ($\mu$) and knowing the effect of the $i$th treatment (factor A) and which of the $j$th nests within the treatment 'the $k$th observation from Block $j$ (factor B) within treatment effect
Variance components
As previously alluded to, it can often be useful to determine the relative contribution (to explaining the unexplained variability) of each of the factors as this provides insights into the variability at each different scales.
These contributions are known as Variance components and are estimates of the added variances due to each of the factors. For consistency with leading texts on this topic, I have included estimated variance components for various balanced nested ANOVA designs in the above table. However, variance components based on a modified version of the maximum likelihood iterative model fitting procedure (REML) is generally recommended as this accommodates both balanced and unbalanced designs.
While there are no numerical differences in the calculations of variance components for fixed and random factors, fixed factors are interpreted very differently and arguably have little biological meaning (other to infer relative contribution). For fixed factors, variance components estimate the variance between the means of the specific populations that are represented by the selected levels of the factor and therefore represent somewhat arbitrary and artificial populations. For random factors, variance components estimate the variance between means of all possible populations that could have been selected and thus represents the true population variance.
Assumptions
Consistent with the hierarchical nature of the design (in which the site means are the appropriate level of replication for the main treatments), it is important that diagnostics associated with each factor reflect this hierarchy. Hence, the replicates for the treatment effects are the site means, and the replicates of the sites are the quadrat means.
- All of the observations are independent - this must be addressed at the design and collection stages. Importantly, to be considered independent replicates, the replicates must be made at the same scale at which the treatment is applied. For example, if the experiment involves subjecting organisms housed in tanks to different water temperatures, then the unit of replication is the individual tanks not the individual organisms in the tanks. The individuals in a tank are strictly not independent with respect to the treatment.
- The response variable (and thus the residuals) should be normally distributed for each sampled populations (combination of factors). Boxplots of each treatment combination are useful for diagnosing major issues with normality.
- The response variable should be equally varied (variance should not be related to mean as these are supposed to be estimated separately) for each combination of treatments. Again, boxplots are useful.
Design balance
In a Bayesian framework, issues of design balance essentially evaporate.
Nested ANOVA in R
Scenario and Data
Imagine we has designed an experiment in which we intend to measure a response ($y$) to one of treatments (three levels; 'a1', 'a2' and 'a3'). The treatments occur at a spatial scale (over an area) that far exceeds the logistical scale of sampling units (it would take too long to sample at the scale at which the treatments were applied). The treatments occurred at the scale of hectares whereas it was only feasible to sample $y$ using 1m quadrats.
Given that the treatments were naturally occurring (such as soil type), it was not possible to have more than five sites of each treatment type, yet prior experience suggested that the sites in which you intended to sample were very uneven and patchy with respect to $y$.
In an attempt to account for this inter-site variability (and thus maximize the power of the test for the effect of treatment, you decided to employ a nested design in which 10 quadrats were randomly located within each of the five replicate sites per three treatments. As this section is mainly about the generation of artificial data (and not specifically about what to do with the data), understanding the actual details are optional and can be safely skipped. Consequently, I have folded (toggled) this section away.
- the number of treatments = 3
- the number of sites per treatment = 5
- the total number of sites = 15
- the sample size per treatment= 5
- the number of quadrats per site = 10
- the mean of the treatments = 40, 70 and 80 respectively
- the variability (standard deviation) between sites of the same treatment = 12
- the variability (standard deviation) between quadrats within sites = 5
> library(plyr) > set.seed(1) > nTreat <- 3 > nSites <- 15 > nSitesPerTreat <- nSites/nTreat > nQuads <- 10 > site.sigma <- 12 > sigma <- 5 > n <- nSites * nQuads > sites <- gl(n = nSites, k = nQuads) > A <- gl(nTreat, nSitesPerTreat * nQuads, n, labels = c("a1", "a2", "a3")) > a.means <- c(40, 70, 80) > ## the site means (treatment effects) are drawn from normal distributions with means of 40, 70 and 80 > ## and standard deviations of 12 > A.effects <- rnorm(nSites, rep(a.means, each = nSitesPerTreat), site.sigma) > # A.effects <- a.means %*% t(model.matrix(~A, > # data.frame(A=gl(nTreat,nSitesPerTreat,nSites))))+rnorm(nSites,0,site.sigma) > Xmat <- model.matrix(~sites - 1) > lin.pred <- Xmat %*% c(A.effects) > ## the quadrat observations (within sites) are drawn from normal distributions with means according to > ## the site means and standard deviations of 5 > y <- rnorm(n, lin.pred, sigma) > data <- data.frame(y = y, A = A, Sites = sites, Quads = 1:length(y)) > head(data) #print out the first six rows of the data set
y A Sites Quads 1 32.26 a1 1 1 2 32.40 a1 1 2 3 37.20 a1 1 3 4 36.59 a1 1 4 5 35.45 a1 1 5 6 37.08 a1 1 6
> library(ggplot2) > ggplot(data, aes(y = y, x = 1)) + geom_boxplot() + facet_grid(. ~ Sites)
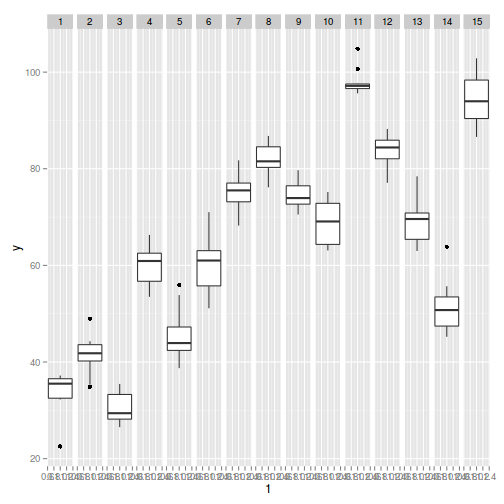
Exploratory data analysis
Normality and Homogeneity of variance
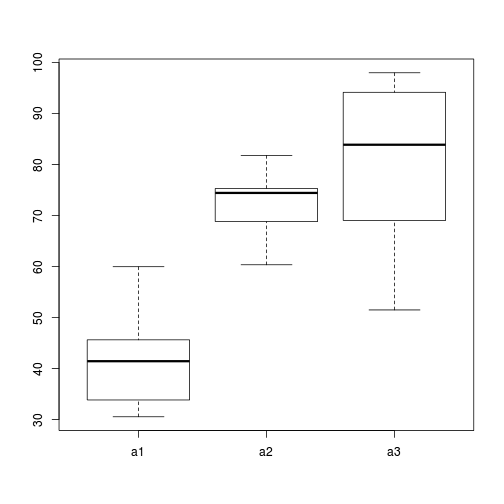
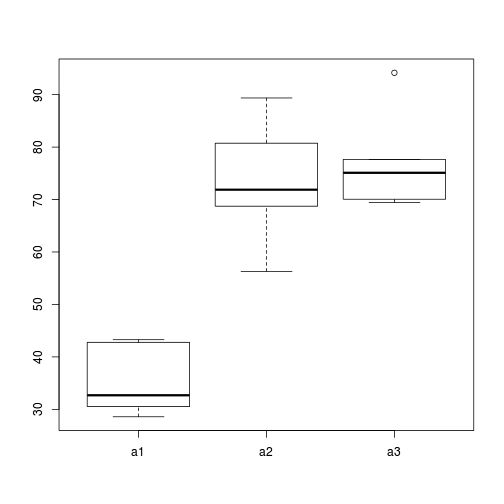
> # Effects of treatment > library(plyr) > boxplot(y ~ A, ddply(data, ~A + Sites, numcolwise(mean, na.rm = T)))
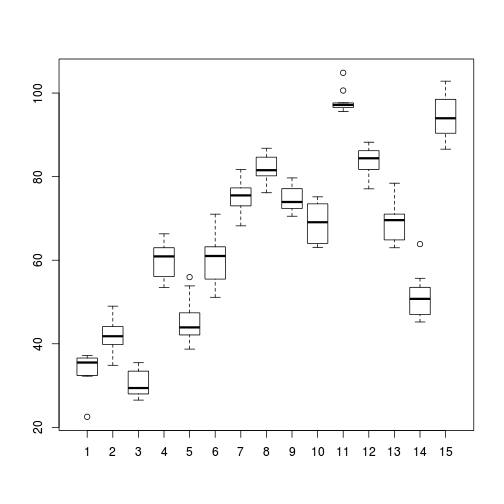
> > # Site effects > boxplot(y ~ Sites, ddply(data, ~A + Sites + Quads, numcolwise(mean, na.rm = T)))
Conclusions:
- there is no evidence that the response variable is consistently non-normal across all populations - each boxplot is approximately symmetrical
- there is no evidence that variance (as estimated by the height of the boxplots) differs between the five populations. . More importantly, there is no evidence of a relationship between mean and variance - the height of boxplots does not increase with increasing position along the y-axis. Hence it there is no evidence of non-homogeneity
- transform the scale of the response variables (to address normality etc). Note transformations should be applied to the entire response variable (not just those populations that are skewed).
JAGS
| Full parameterization | Matrix parameterization | Heirarchical parameterization |
|---|---|---|
| $$ \begin{array}{rcl} y_{ijk}&\sim&\mathcal{N}(\mu_{ij},\sigma^2)\\ \mu_{ij} &=& \alpha_0 + \alpha_{i} + \beta_{j(i)}\\ \beta_{i{j}}&\sim&\mathcal{N}(0,\sigma_{B}^2)\\ \alpha_0, \alpha_i&\sim&\mathcal{N}(0,100000)\\ \sigma^2&\sim&\mathcal{U}(0,100)\\ \sigma_{B}^2&\sim&\mathcal{U}(0,100)\\ \end{array} $$ | $$ \begin{array}{rcl} y_{ijk}&\sim&\mathcal{N}(\mu_{ij},\sigma^2)\\ \mu_{ij} &=& \alpha\mathbf{X} + \beta_{j(i)}\\ \beta_{i{j}}&\sim&\mathcal{N}(0,\sigma_{B}^2)\\ \alpha&\sim&\mathcal{MVN}(0,100000)\\ \sigma^2&\sim&\mathcal{U}(0,100)\\ \sigma_{B}^2&\sim&\mathcal{U}(0,100)\\ \end{array} $$ | $$ \begin{array}{rcl} y_{ij}&\sim &\mathcal{N}(\beta_{j(i)}, \sigma^2)\\ \beta_{j(i)}&\sim&\mathcal{N}(\mu_{i},\sigma_{B}^2)\\ \mu_{i}&=&\alpha\mathbf{X}\\ \alpha_i&\sim&\mathcal{N}(0, 1000000)\\ \sigma^2&\sim&\mathcal{U}(0,100)\\ \sigma_{B}^2&\sim&\mathcal{U}(0,100) \end{array} $$ |
The full parameterization, shows the effects parameterization in which there is an intercept ($\alpha_0$) and two treatment effects ($\alpha_i$, where $i$ is 1,2).
The matrix parameterization is a compressed notation, In this parameterization, there are three alpha parameters (one representing the mean of treatment a1, and the other two representing the treatment effects (differences between a2 and a1 and a3 and a1). In generating priors for each of these three alpha parameters, we could loop through each and define a non-informative normal prior to each (as in the Full parameterization version). However, it turns out that it is more efficient (in terms of mixing and thus the number of necessary iterations) to define the priors from a multivariate normal distribution. This has as many means as there are parameters to estimate (3) and a 3x3 matrix of zeros and 100 in the diagonals. $$ \mu\sim\left[ \begin{array}{c} 0\\ 0\\ 0\\ \end{array} \right], \hspace{2em} \sigma^2\sim\left[ \begin{array}{ccc} 1000000&0&0\\ 0&1000000&0\\ 0&0&1000000\\ \end{array} \right] $$
In the Heirarchical parameterization, we are indicating two residual layers - one representing the variability in the observed data between individual observations (within sites) and the second representing the variability between site means (within the three treatments).
Full effects parameterization
> modelString=" model { #Likelihood for (i in 1:n) { y[i]~dnorm(mu[i],tau) mu[i] <- alpha0 + alpha[A[i]] + beta[site[i]] } #Priors alpha0 ~ dnorm(0, 1.0E-6) alpha[1] <- 0 for (i in 2:nA) { alpha[i] ~ dnorm(0, 1.0E-6) #prior } for (i in 1:nSite) { beta[i] ~ dnorm(0, tau.B) #prior } sigma ~ dunif(0, 100) tau <- 1 / (sigma * sigma) sigma.B ~ dunif(0, 100) tau.B <- 1 / (sigma.B * sigma.B) } " > writeLines(modelString,con="../downloads/BUGSscripts/tut9.7bS6.1.f.txt")
> data.list <- with(data, + list(y=y, + site=as.numeric(Sites), + A=as.numeric(A), + n=nrow(data), + nSite=length(levels(Sites)), + nA = length(levels(A)) + ) + ) > > params <- c("alpha0","alpha","sigma","sigma.B") > adaptSteps = 1000 > burnInSteps = 3000 > nChains = 3 > numSavedSteps = 3000 > thinSteps = 10 > nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) > > library(R2jags) > rnorm(1)
[1] -0.5429
> jags.effects.f.time <- system.time( + data.r2jags.f <- jags(data=data.list, + inits=NULL, + parameters.to.save=params, + model.file="../downloads/BUGSscripts/tut9.7bS6.1.f.txt", + n.chains=3, + n.iter=nIter, + n.burnin=burnInSteps, + n.thin=thinSteps + ) + )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 497 Initializing model
> jags.effects.f.time
user system elapsed 3.892 0.004 3.926
> print(data.r2jags.f)
Inference for Bugs model at "../downloads/BUGSscripts/tut9.7bS6.1.f.txt", fit using jags,
3 chains, each with 13000 iterations (first 3000 discarded), n.thin = 10
n.sims = 3000 iterations saved
mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff
alpha[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1
alpha[2] 29.757 10.126 9.415 23.489 29.744 36.048 50.397 1.001 3000
alpha[3] 36.827 10.062 16.002 30.585 37.001 43.287 56.551 1.004 630
alpha0 42.372 7.056 28.368 38.069 42.409 46.828 56.357 1.003 1200
sigma 4.415 0.272 3.928 4.221 4.402 4.597 4.992 1.001 3000
sigma.B 15.294 3.613 10.130 12.787 14.727 17.132 24.044 1.001 3000
deviance 869.502 5.926 859.939 865.291 868.786 873.034 883.015 1.001 2600
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 17.6 and DIC = 887.1
DIC is an estimate of expected predictive error (lower deviance is better).
> data.mcmc.list.f <- as.mcmc(data.r2jags.f)
[1] "alpha[1]" "alpha[2]" "alpha[3]" "alpha0" "deviance" "sigma" "sigma.B"
Matrix parameterization
> modelString=" model { #Likelihood for (i in 1:n) { y[i]~dnorm(mu[i],tau) mu[i] <- inprod(alpha[],X[i,]) + beta[site[i]] } #Priors alpha ~ dmnorm(a0,A0) for (i in 1:nSite) { beta[i] ~ dnorm(0, tau.B) #prior } sigma ~ dunif(0, 100) tau <- 1 / (sigma * sigma) sigma.B ~ dunif(0, 100) tau.B <- 1 / (sigma.B * sigma.B) } " > writeLines(modelString,con="../downloads/BUGSscripts/tut9.7bS6.1.m.txt")
> A.Xmat <- model.matrix(~A,data) > data.list <- with(data, + list(y=y, + site=as.numeric(Sites), + X=A.Xmat, + n=nrow(data), + nSite=length(levels(Sites)), + nA = ncol(A.Xmat), + a0=rep(0,3), A0=diag(0,3) + ) + ) > > params <- c("alpha","sigma","sigma.B") > adaptSteps = 1000 > burnInSteps = 3000 > nChains = 3 > numSavedSteps = 3000 > thinSteps = 10 > nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) > > library(R2jags) > rnorm(1)
[1] -0.3613
> jags.effects.m.time <- system.time( + data.r2jags.m <- jags(data=data.list, + inits=NULL, + parameters.to.save=params, + model.file="../downloads/BUGSscripts/tut9.7bS6.1.m.txt", + n.chains=3, + n.iter=nIter, + n.burnin=burnInSteps, + n.thin=thinSteps + ) + )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 959 Initializing model
> jags.effects.m.time
user system elapsed 4.096 0.000 4.101
> print(data.r2jags.m)
Inference for Bugs model at "../downloads/BUGSscripts/tut9.7bS6.1.m.txt", fit using jags,
3 chains, each with 13000 iterations (first 3000 discarded), n.thin = 10
n.sims = 3000 iterations saved
mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff
alpha[1] 42.392 7.125 28.626 37.676 42.452 46.841 56.358 1.001 3000
alpha[2] 29.560 10.022 10.006 23.317 29.663 35.702 49.278 1.001 3000
alpha[3] 36.996 10.012 16.417 30.832 37.155 43.348 55.847 1.001 3000
sigma 4.416 0.272 3.913 4.223 4.402 4.595 4.978 1.001 3000
sigma.B 15.355 3.696 10.122 12.765 14.709 17.193 24.493 1.001 3000
deviance 869.704 6.050 859.995 865.340 868.843 873.503 883.052 1.001 3000
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 18.3 and DIC = 888.0
DIC is an estimate of expected predictive error (lower deviance is better).
> data.mcmc.list.m <- as.mcmc(data.r2jags.m) > Data.mcmc.list.m <- data.mcmc.list.m
Hierarchical parameterization
> modelString=" model { #Likelihood (esimating site means (gamma.site) for (i in 1:n) { y[i]~dnorm(quad.means[i],tau) quad.means[i] <- gamma.site[site[i]] } for (i in 1:s) { gamma.site[i] ~ dnorm(site.means[i], tau.site) site.means[i] <- inprod(beta[],A.Xmat[i,]) } #Priors for (i in 1:a) { beta[i] ~ dnorm(0, 1.0E-6) #prior } sigma ~ dunif(0, 100) tau <- 1 / (sigma * sigma) sigma.site ~ dunif(0, 100) tau.site <- 1 / (sigma.site * sigma.site) } " > writeLines(modelString,con="../downloads/BUGSscripts/tut9.7bS6.1.txt")
> A.Xmat <- model.matrix(~A,ddply(data,~Sites,catcolwise(unique))) > data.list <- with(data, + list(y=y, + site=Sites, + A.Xmat= A.Xmat, + n=nrow(data), + s=length(levels(Sites)), + a = ncol(A.Xmat) + ) + ) > > params <- c("beta","sigma","sigma.site") > adaptSteps = 1000 > burnInSteps = 3000 > nChains = 3 > numSavedSteps = 3000 > thinSteps = 10 > nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) > > library(R2jags) > rnorm(1)
[1] -0.1138
> jags.effects.simple.time <- system.time( + data.r2jags <- jags(data=data.list, + inits=NULL, + parameters.to.save=params, + model.file="../downloads/BUGSscripts/tut9.7bS6.1.txt", + n.chains=3, + n.iter=nIter, + n.burnin=burnInSteps, + n.thin=thinSteps + ) + )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 395 Initializing model
> print(data.r2jags)
Inference for Bugs model at "../downloads/BUGSscripts/tut9.7bS6.1.txt", fit using jags,
3 chains, each with 13000 iterations (first 3000 discarded), n.thin = 10
n.sims = 3000 iterations saved
mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff
beta[1] 42.172 6.952 28.859 37.707 42.124 46.416 56.249 1.001 3000
beta[2] 29.947 9.819 10.441 23.568 29.956 36.217 49.381 1.001 3000
beta[3] 37.051 9.961 16.859 30.916 37.298 43.406 56.494 1.001 3000
sigma 4.405 0.271 3.931 4.217 4.386 4.589 4.976 1.001 3000
sigma.site 15.402 3.756 10.034 12.760 14.741 17.289 24.589 1.001 3000
deviance 869.480 6.000 859.690 865.195 868.708 873.196 882.851 1.001 3000
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 18.0 and DIC = 887.5
DIC is an estimate of expected predictive error (lower deviance is better).
> data.mcmc.list <- as.mcmc(data.r2jags)
> modelString=" model { #Likelihood (esimating site means (gamma.site) for (i in 1:n) { y[i]~dnorm(quad.means[i],tau) quad.means[i] <- gamma.site[site[i]] y.err[i]<- quad.means[i]-y[i] } for (i in 1:s) { gamma.site[i] ~ dnorm(site.means[i], tau.site) site.means[i] <- inprod(beta[],A.Xmat[i,]) site.err[i] <- site.means[i] - gamma.site[i] } #Priors for (i in 1:a) { beta[i] ~ dnorm(0, 1.0E-6) #prior } sigma ~ dunif(0, 100) tau <- 1 / (sigma * sigma) sigma.site ~ dunif(0, 100) tau.site <- 1 / (sigma.site * sigma.site) sd.y <- sd(y.err) sd.site <- sd(site.err) sd.A <- sd(beta) } " > writeLines(modelString,con="../downloads/BUGSscripts/tut9.7bS6.1SD.txt")
> A.Xmat <- model.matrix(~A,ddply(data,~Sites,catcolwise(unique))) > data.list <- with(data, + list(y=y, + site=Sites, + A.Xmat= A.Xmat, + n=nrow(data), + s=length(levels(Sites)), + a = ncol(A.Xmat) + ) + ) > > params <- c("beta","sigma","sd.y",'sd.site','sd.A','sigma','sigma.site') > adaptSteps = 1000 > burnInSteps = 3000 > nChains = 3 > numSavedSteps = 3000 > thinSteps = 10 > nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) > > library(R2jags) > rnorm(1)
[1] -1.029
> jags.SD.time <- system.time( + data.r2jagsSD <- jags(data=data.list, + inits=NULL, + parameters.to.save=params, + model.file="../downloads/BUGSscripts/tut9.7bS6.1SD.txt", + n.chains=3, + n.iter=nIter, + n.burnin=burnInSteps, + n.thin=thinSteps + ) + )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 565 Initializing model
> print(data.r2jagsSD)
Inference for Bugs model at "../downloads/BUGSscripts/tut9.7bS6.1SD.txt", fit using jags,
3 chains, each with 13000 iterations (first 3000 discarded), n.thin = 10
n.sims = 3000 iterations saved
mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff
beta[1] 42.137 7.441 27.159 37.585 42.077 46.869 56.838 1.007 1800
beta[2] 29.830 10.136 9.088 23.728 29.813 35.965 49.683 1.004 3000
beta[3] 37.283 10.161 16.716 31.085 37.398 43.308 57.374 1.001 3000
sd.A 8.434 5.007 1.523 4.835 7.618 10.968 20.088 1.004 600
sd.site 13.456 1.535 11.828 12.512 13.062 13.891 17.371 1.007 420
sd.y 4.359 0.082 4.228 4.299 4.348 4.408 4.547 1.002 1800
sigma 4.410 0.268 3.909 4.220 4.398 4.591 4.972 1.001 3000
sigma.site 15.381 3.781 10.083 12.748 14.744 17.261 24.709 1.001 3000
deviance 869.274 5.901 859.676 865.005 868.651 872.882 882.650 1.002 1500
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 17.4 and DIC = 886.7
DIC is an estimate of expected predictive error (lower deviance is better).
> data.mcmc.listSD <- as.mcmc(data.r2jagsSD)
Rstan
Cell means parameterization
> rstanString=" data{ int n; int nA; int nB; vector [n] y; int A[n]; int B[n]; } parameters{ real alpha[nA]; real<lower=0> sigma; vector [nB] beta; real<lower=0> sigma_B; } model{ real mu[n]; // Priors alpha ~ normal( 0 , 100 ); beta ~ normal( 0 , sigma_B ); sigma_B ~ uniform( 0 , 100 ); sigma ~ uniform( 0 , 100 ); for ( i in 1:n ) { mu[i] <- alpha[A[i]] + beta[B[i]]; } y ~ normal( mu , sigma ); } "
> data.list <- with(data, list(y=y, A=as.numeric(A), B=as.numeric(Sites), n=nrow(data), nB=length(levels(Sites)),nA=length(levels(A)))) > burnInSteps = 3000 > nChains = 3 > numSavedSteps = 3000 > thinSteps = 10 > nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) > library(rstan) > rstan.c.time <- system.time( + data.rstan.c <- stan(data=data.list, + model_code=rstanString, + pars=c('alpha','sigma','sigma_B'), + chains=nChains, + iter=nIter, + warmup=burnInSteps, + thin=thinSteps, + save_dso=TRUE + ) + )
TRANSLATING MODEL 'rstanString' FROM Stan CODE TO C++ CODE NOW.
COMPILING THE C++ CODE FOR MODEL 'rstanString' NOW.
SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 1).
Iteration: 1 / 13000 [ 0%] (Warmup)
Iteration: 1300 / 13000 [ 10%] (Warmup)
Iteration: 2600 / 13000 [ 20%] (Warmup)
Iteration: 3900 / 13000 [ 30%] (Sampling)
Iteration: 5200 / 13000 [ 40%] (Sampling)
Iteration: 6500 / 13000 [ 50%] (Sampling)
Iteration: 7800 / 13000 [ 60%] (Sampling)
Iteration: 9100 / 13000 [ 70%] (Sampling)
Iteration: 10400 / 13000 [ 80%] (Sampling)
Iteration: 11700 / 13000 [ 90%] (Sampling)
Iteration: 13000 / 13000 [100%] (Sampling)
Elapsed Time: 0.99 seconds (Warm-up)
4 seconds (Sampling)
4.99 seconds (Total)
SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 2).
Iteration: 1 / 13000 [ 0%] (Warmup)
Iteration: 1300 / 13000 [ 10%] (Warmup)
Iteration: 2600 / 13000 [ 20%] (Warmup)
Iteration: 3900 / 13000 [ 30%] (Sampling)
Iteration: 5200 / 13000 [ 40%] (Sampling)
Iteration: 6500 / 13000 [ 50%] (Sampling)
Iteration: 7800 / 13000 [ 60%] (Sampling)
Iteration: 9100 / 13000 [ 70%] (Sampling)
Iteration: 10400 / 13000 [ 80%] (Sampling)
Iteration: 11700 / 13000 [ 90%] (Sampling)
Iteration: 13000 / 13000 [100%] (Sampling)
Elapsed Time: 1.22 seconds (Warm-up)
4.16 seconds (Sampling)
5.38 seconds (Total)
SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 3).
Iteration: 1 / 13000 [ 0%] (Warmup)
Iteration: 1300 / 13000 [ 10%] (Warmup)
Iteration: 2600 / 13000 [ 20%] (Warmup)
Iteration: 3900 / 13000 [ 30%] (Sampling)
Iteration: 5200 / 13000 [ 40%] (Sampling)
Iteration: 6500 / 13000 [ 50%] (Sampling)
Iteration: 7800 / 13000 [ 60%] (Sampling)
Iteration: 9100 / 13000 [ 70%] (Sampling)
Iteration: 10400 / 13000 [ 80%] (Sampling)
Iteration: 11700 / 13000 [ 90%] (Sampling)
Iteration: 13000 / 13000 [100%] (Sampling)
Elapsed Time: 1.09 seconds (Warm-up)
3.79 seconds (Sampling)
4.88 seconds (Total)
> print(data.rstan.c)
Inference for Stan model: rstanString.
3 chains, each with iter=13000; warmup=3000; thin=10;
post-warmup draws per chain=1000, total post-warmup draws=3000.
mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat
alpha[1] 41.9 0.1 7.2 27.5 37.5 42.2 46.5 56.3 2616 1
alpha[2] 71.8 0.1 7.1 57.6 67.4 71.9 76.2 85.8 2322 1
alpha[3] 78.8 0.1 7.0 64.2 74.6 78.9 83.2 92.1 2749 1
sigma 4.4 0.0 0.3 3.9 4.2 4.4 4.6 5.0 3000 1
sigma_B 15.3 0.1 3.7 10.0 12.7 14.6 17.2 24.5 2384 1
lp__ -340.9 0.1 3.4 -348.3 -343.0 -340.5 -338.4 -335.3 2934 1
Samples were drawn using NUTS(diag_e) at Thu Mar 13 18:27:13 2014.
For each parameter, n_eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor on split chains (at
convergence, Rhat=1).
> > > > data.rstan.c.df <-as.data.frame(extract(data.rstan.c)) > head(data.rstan.c.df)
alpha.1 alpha.2 alpha.3 sigma sigma_B lp__ 1 55.85 81.35 77.26 4.607 13.382 -343.8 2 41.60 59.31 80.78 4.778 15.285 -347.9 3 45.26 73.41 73.49 4.500 15.735 -337.7 4 49.88 76.52 71.62 4.785 22.865 -351.2 5 37.07 69.37 83.65 4.451 9.727 -343.8 6 48.24 62.19 70.40 4.411 13.340 -349.0
> > data.mcmc.c<-rstan:::as.mcmc.list.stanfit(data.rstan.c) > > > plyr:::adply(as.matrix(data.rstan.c.df),2,MCMCsum)
X1 Median X0. X25. X50. X75. X100. lower upper lower.1 upper.1 1 alpha.1 42.16 1.935 37.516 42.16 46.51 68.27 26.694 55.200 36.136 49.536 2 alpha.2 71.91 30.043 67.384 71.91 76.15 115.11 58.013 86.164 65.380 78.547 3 alpha.3 78.86 41.298 74.612 78.86 83.20 104.32 65.060 92.759 72.504 85.283 4 sigma 4.40 3.631 4.236 4.40 4.59 5.34 3.913 4.973 4.156 4.688 5 sigma_B 14.62 7.302 12.661 14.62 17.23 42.28 9.444 22.445 11.030 17.319 6 lp__ -340.53 -355.277 -343.037 -340.53 -338.39 -332.32 -347.638 -334.809 -343.626 -337.055
Full effects parameterization
> rstanString=" data{ int n; int nB; vector [n] y; int A2[n]; int A3[n]; int B[n]; } parameters{ real alpha0; real alpha2; real alpha3; real<lower=0> sigma; vector [nB] beta; real<lower=0> sigma_B; } model{ real mu[n]; // Priors alpha0 ~ normal( 0 , 100 ); alpha2 ~ normal( 0 , 100 ); alpha3 ~ normal( 0 , 100 ); beta ~ normal( 0 , sigma_B ); sigma_B ~ uniform( 0 , 100 ); sigma ~ uniform( 0 , 100 ); for ( i in 1:n ) { mu[i] <- alpha0 + alpha2*A2[i] + alpha3*A3[i] + beta[B[i]]; } y ~ normal( mu , sigma ); } "
> A2 <- ifelse(data$A=='a2',1,0) > A3 <- ifelse(data$A=='a3',1,0) > data.list <- with(data, list(y=y, A2=A2, A3=A3, B=as.numeric(Sites), n=nrow(data), nB=length(levels(Sites)))) > burnInSteps = 3000 > nChains = 3 > numSavedSteps = 3000 > thinSteps = 10 > nIter = burnInSteps + ceiling((numSavedSteps * thinSteps)/nChains) > library(rstan) > rstan.f.time <- system.time( + data.rstan.f <- stan(data=data.list, + model_code=rstanString, + pars=c('alpha0','alpha2','alpha3','sigma','sigma_B'), + chains=nChains, + iter=nIter, + warmup=burnInSteps, + thin=thinSteps, + save_dso=TRUE + ) + )
TRANSLATING MODEL 'rstanString' FROM Stan CODE TO C++ CODE NOW.
COMPILING THE C++ CODE FOR MODEL 'rstanString' NOW.
SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 1).
Iteration: 1 / 13000 [ 0%] (Warmup)
Iteration: 1300 / 13000 [ 10%] (Warmup)
Iteration: 2600 / 13000 [ 20%] (Warmup)
Iteration: 3900 / 13000 [ 30%] (Sampling)
Iteration: 5200 / 13000 [ 40%] (Sampling)
Iteration: 6500 / 13000 [ 50%] (Sampling)
Iteration: 7800 / 13000 [ 60%] (Sampling)
Iteration: 9100 / 13000 [ 70%] (Sampling)
Iteration: 10400 / 13000 [ 80%] (Sampling)
Iteration: 11700 / 13000 [ 90%] (Sampling)
Iteration: 13000 / 13000 [100%] (Sampling)
Elapsed Time: 2.97 seconds (Warm-up)
12.45 seconds (Sampling)
15.42 seconds (Total)
SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 2).
Iteration: 1 / 13000 [ 0%] (Warmup)
Iteration: 1300 / 13000 [ 10%] (Warmup)
Iteration: 2600 / 13000 [ 20%] (Warmup)
Iteration: 3900 / 13000 [ 30%] (Sampling)
Iteration: 5200 / 13000 [ 40%] (Sampling)
Iteration: 6500 / 13000 [ 50%] (Sampling)
Iteration: 7800 / 13000 [ 60%] (Sampling)
Iteration: 9100 / 13000 [ 70%] (Sampling)
Iteration: 10400 / 13000 [ 80%] (Sampling)
Iteration: 11700 / 13000 [ 90%] (Sampling)
Iteration: 13000 / 13000 [100%] (Sampling)
Elapsed Time: 3.45 seconds (Warm-up)
14.69 seconds (Sampling)
18.14 seconds (Total)
SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 3).
Iteration: 1 / 13000 [ 0%] (Warmup)
Iteration: 1300 / 13000 [ 10%] (Warmup)
Iteration: 2600 / 13000 [ 20%] (Warmup)
Iteration: 3900 / 13000 [ 30%] (Sampling)
Iteration: 5200 / 13000 [ 40%] (Sampling)
Iteration: 6500 / 13000 [ 50%] (Sampling)
Iteration: 7800 / 13000 [ 60%] (Sampling)
Iteration: 9100 / 13000 [ 70%] (Sampling)
Iteration: 10400 / 13000 [ 80%] (Sampling)
Iteration: 11700 / 13000 [ 90%] (Sampling)
Iteration: 13000 / 13000 [100%] (Sampling)
Elapsed Time: 2.89 seconds (Warm-up)
13.94 seconds (Sampling)
16.83 seconds (Total)
> print(data.rstan.f)
Inference for Stan model: rstanString.
3 chains, each with iter=13000; warmup=3000; thin=10;
post-warmup draws per chain=1000, total post-warmup draws=3000.
mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat
alpha0 42.1 0.1 7.0 28.0 37.6 42.1 46.4 55.9 3000 1
alpha2 29.8 0.2 9.9 10.5 23.4 30.0 36.3 49.8 3000 1
alpha3 37.1 0.2 9.9 17.4 30.7 37.2 43.3 57.0 2953 1
sigma 4.4 0.0 0.3 3.9 4.2 4.4 4.6 5.0 3000 1
sigma_B 15.4 0.1 3.7 10.1 12.8 14.8 17.3 24.0 2674 1
lp__ -340.5 0.1 3.4 -348.2 -342.6 -340.1 -338.1 -334.9 2603 1
Samples were drawn using NUTS(diag_e) at Thu Mar 13 18:35:44 2014.
For each parameter, n_eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor on split chains (at
convergence, Rhat=1).
> > > > data.rstan.f.df <-as.data.frame(extract(data.rstan.f)) > head(data.rstan.f.df)
alpha0 alpha2 alpha3 sigma sigma_B lp__ 1 50.04 29.56 34.25 4.586 15.12 -335.6 2 30.58 47.51 51.30 4.292 10.64 -343.1 3 38.09 35.59 46.83 4.728 12.33 -343.3 4 45.31 32.96 30.53 4.429 16.41 -337.9 5 39.15 29.35 34.07 4.161 14.22 -338.4 6 35.63 30.33 41.81 4.373 11.24 -336.8
> > data.mcmc.f<-rstan:::as.mcmc.list.stanfit(data.rstan.f) > > > plyr:::adply(as.matrix(data.rstan.f.df),2,MCMCsum)
X1 Median X0. X25. X50. X75. X100. lower upper lower.1 upper.1 1 alpha0 42.100 6.560 37.58 42.100 46.449 71.967 28.583 56.321 35.58 48.805 2 alpha2 29.990 -11.516 23.39 29.990 36.287 67.775 10.433 49.769 20.11 38.919 3 alpha3 37.233 -4.153 30.70 37.233 43.270 80.874 17.553 57.083 28.31 47.125 4 sigma 4.406 3.504 4.23 4.406 4.583 5.432 3.938 5.004 4.14 4.652 5 sigma_B 14.780 8.260 12.82 14.780 17.261 55.029 9.404 22.292 11.23 17.471 6 lp__ -340.081 -359.684 -342.58 -340.081 -338.077 -332.487 -347.214 -334.170 -342.86 -336.394
Matrix parameterization
> rstanString=" data{ int n; int nB; int nA; vector [n] y; matrix [n,nA] X; int B[n]; vector [nA] a0; matrix [nA,nA] A0; } parameters{ vector [nA] alpha; real<lower=0> sigma; vector [nB] beta; real<lower=0> sigma_B; } model{ real mu[n]; // Priors //alpha ~ normal( 0 , 100 ); alpha ~ multi_normal(a0,A0); beta ~ normal( 0 , sigma_B ); sigma_B ~ uniform( 0 , 100 ); sigma ~ uniform( 0 , 100 ); for ( i in 1:n ) { mu[i] <- dot_product(X[i],alpha) + beta[B[i]]; } y ~ normal( mu , sigma ); } "
> X <- model.matrix(~A, data) > nA <- ncol(X) > data.list <- with(data, list(y=y, X=X, B=as.numeric(Sites), n=nrow(data), nB=length(levels(Sites)), nA=nA, + a0=rep(0,nA), A0=diag(100000,nA))) > burnInSteps = 3000 > nChains = 3 > numSavedSteps = 3000 > thinSteps = 10 > nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) > library(rstan) > rstan.m.time <- system.time( + data.rstan.m <- stan(data=data.list, + model_code=rstanString, + pars=c('alpha','sigma','sigma_B'), + chains=nChains, + iter=nIter, + warmup=burnInSteps, + thin=thinSteps, + save_dso=TRUE + ) + )
TRANSLATING MODEL 'rstanString' FROM Stan CODE TO C++ CODE NOW.
COMPILING THE C++ CODE FOR MODEL 'rstanString' NOW.
SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 1).
Iteration: 1 / 13000 [ 0%] (Warmup)
Iteration: 1300 / 13000 [ 10%] (Warmup)
Iteration: 2600 / 13000 [ 20%] (Warmup)
Iteration: 3900 / 13000 [ 30%] (Sampling)
Iteration: 5200 / 13000 [ 40%] (Sampling)
Iteration: 6500 / 13000 [ 50%] (Sampling)
Iteration: 7800 / 13000 [ 60%] (Sampling)
Iteration: 9100 / 13000 [ 70%] (Sampling)
Iteration: 10400 / 13000 [ 80%] (Sampling)
Iteration: 11700 / 13000 [ 90%] (Sampling)
Iteration: 13000 / 13000 [100%] (Sampling)
Elapsed Time: 5.26 seconds (Warm-up)
19.14 seconds (Sampling)
24.4 seconds (Total)
SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 2).
Iteration: 1 / 13000 [ 0%] (Warmup)
Iteration: 1300 / 13000 [ 10%] (Warmup)
Iteration: 2600 / 13000 [ 20%] (Warmup)
Iteration: 3900 / 13000 [ 30%] (Sampling)
Iteration: 5200 / 13000 [ 40%] (Sampling)
Iteration: 6500 / 13000 [ 50%] (Sampling)
Iteration: 7800 / 13000 [ 60%] (Sampling)
Iteration: 9100 / 13000 [ 70%] (Sampling)
Iteration: 10400 / 13000 [ 80%] (Sampling)
Iteration: 11700 / 13000 [ 90%] (Sampling)
Iteration: 13000 / 13000 [100%] (Sampling)
Elapsed Time: 5.25 seconds (Warm-up)
29.73 seconds (Sampling)
34.98 seconds (Total)
SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 3).
Iteration: 1 / 13000 [ 0%] (Warmup)
Iteration: 1300 / 13000 [ 10%] (Warmup)
Iteration: 2600 / 13000 [ 20%] (Warmup)
Iteration: 3900 / 13000 [ 30%] (Sampling)
Iteration: 5200 / 13000 [ 40%] (Sampling)
Iteration: 6500 / 13000 [ 50%] (Sampling)
Iteration: 7800 / 13000 [ 60%] (Sampling)
Iteration: 9100 / 13000 [ 70%] (Sampling)
Iteration: 10400 / 13000 [ 80%] (Sampling)
Iteration: 11700 / 13000 [ 90%] (Sampling)
Iteration: 13000 / 13000 [100%] (Sampling)
Elapsed Time: 5.63 seconds (Warm-up)
20.57 seconds (Sampling)
26.2 seconds (Total)
> print(data.rstan.m)
Inference for Stan model: rstanString.
3 chains, each with iter=13000; warmup=3000; thin=10;
post-warmup draws per chain=1000, total post-warmup draws=3000.
mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat
alpha[1] 42.2 0.1 7.1 27.8 37.7 42.3 46.8 56.0 2886 1
alpha[2] 29.9 0.2 10.0 10.8 23.4 29.7 35.9 50.5 2847 1
alpha[3] 37.1 0.2 10.2 16.9 30.7 37.1 43.4 57.8 2619 1
sigma 4.4 0.0 0.3 3.9 4.2 4.4 4.6 5.0 3000 1
sigma_B 15.4 0.1 3.8 10.0 12.7 14.8 17.3 24.1 2603 1
lp__ -340.4 0.1 3.5 -348.1 -342.5 -340.1 -337.9 -334.8 2802 1
Samples were drawn using NUTS(diag_e) at Fri Mar 14 12:11:04 2014.
For each parameter, n_eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor on split chains (at
convergence, Rhat=1).
> > data.rstan.m.df <-as.data.frame(extract(data.rstan.m)) > head(data.rstan.m.df)
alpha.1 alpha.2 alpha.3 sigma sigma_B lp__ 1 50.14 19.46 29.41 4.806 13.44 -340.0 2 41.98 31.19 33.78 4.010 13.82 -336.8 3 45.76 34.30 30.29 4.200 15.04 -335.4 4 37.32 38.25 37.28 4.274 13.19 -341.2 5 33.74 34.28 53.22 4.236 17.93 -342.8 6 56.95 16.36 36.67 4.406 15.01 -343.8
> > data.mcmc.m<-rstan:::as.mcmc.list.stanfit(data.rstan.m) > > plyr:::adply(as.matrix(data.rstan.m.df),2,MCMCsum)
X1 Median X0. X25. X50. X75. X100. lower upper lower.1 upper.1 1 alpha.1 42.303 8.685 37.723 42.303 46.784 69.630 28.761 56.777 36.266 49.570 2 alpha.2 29.678 -15.254 23.424 29.678 35.903 72.365 9.756 49.231 20.322 38.784 3 alpha.3 37.055 -8.139 30.657 37.055 43.424 90.338 14.785 55.643 26.635 45.559 4 sigma 4.399 3.561 4.223 4.399 4.599 5.551 3.872 4.926 4.131 4.674 5 sigma_B 14.760 8.405 12.739 14.760 17.276 56.891 9.403 22.740 11.174 17.580 6 lp__ -340.066 -357.700 -342.529 -340.066 -337.927 -331.699 -347.418 -334.411 -342.985 -336.462
Heirarchical parameterization
> rstanString=" data{ int n; int nA; int nSites; vector [n] y; matrix [nSites,nA] X; matrix [n,nSites] Z; } parameters{ vector[nA] beta; vector[nSites] gamma; real<lower=0> sigma; real<lower=0> sigma_S; } model{ vector [n] mu_site; vector [nSites] mu; // Priors beta ~ normal( 0 , 1000 ); gamma ~ normal( 0 , 1000 ); sigma ~ uniform( 0 , 100 ); sigma_S~ uniform( 0 , 100 ); mu_site <- Z*gamma; y ~ normal( mu_site , sigma ); mu <- X*beta; gamma ~ normal(mu, sigma_S); } "
> dt.A <- ddply(data,~Sites,catcolwise(unique)) > X<-model.matrix(~A, dt.A) > Z<-model.matrix(~Sites-1, data) > data.list <- list(y=data$y, X=X, Z=Z, n=nrow(data), nSites=nrow(X),nA=ncol(X)) > burnInSteps = 2000 > nChains = 3 > numSavedSteps = 3000 > thinSteps = 10 > nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) > library(rstan) > rstan.time <- system.time( + data.rstan <- stan(data=data.list, + model_code=rstanString, + pars=c('beta','sigma','sigma_S'), + chains=nChains, + iter=nIter, + warmup=burnInSteps, + thin=thinSteps, + save_dso=TRUE + ) + )
TRANSLATING MODEL 'rstanString' FROM Stan CODE TO C++ CODE NOW.
COMPILING THE C++ CODE FOR MODEL 'rstanString' NOW.
SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 1).
Iteration: 1 / 12000 [ 0%] (Warmup)
Iteration: 1200 / 12000 [ 10%] (Warmup)
Iteration: 2400 / 12000 [ 20%] (Sampling)
Iteration: 3600 / 12000 [ 30%] (Sampling)
Iteration: 4800 / 12000 [ 40%] (Sampling)
Iteration: 6000 / 12000 [ 50%] (Sampling)
Iteration: 7200 / 12000 [ 60%] (Sampling)
Iteration: 8400 / 12000 [ 70%] (Sampling)
Iteration: 9600 / 12000 [ 80%] (Sampling)
Iteration: 10800 / 12000 [ 90%] (Sampling)
Iteration: 12000 / 12000 [100%] (Sampling)
Elapsed Time: 0.99 seconds (Warm-up)
2.23 seconds (Sampling)
3.22 seconds (Total)
SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 2).
Iteration: 1 / 12000 [ 0%] (Warmup)
Iteration: 1200 / 12000 [ 10%] (Warmup)
Iteration: 2400 / 12000 [ 20%] (Sampling)
Iteration: 3600 / 12000 [ 30%] (Sampling)
Iteration: 4800 / 12000 [ 40%] (Sampling)
Iteration: 6000 / 12000 [ 50%] (Sampling)
Iteration: 7200 / 12000 [ 60%] (Sampling)
Iteration: 8400 / 12000 [ 70%] (Sampling)
Iteration: 9600 / 12000 [ 80%] (Sampling)
Iteration: 10800 / 12000 [ 90%] (Sampling)
Iteration: 12000 / 12000 [100%] (Sampling)
Elapsed Time: 1.02 seconds (Warm-up)
3 seconds (Sampling)
4.02 seconds (Total)
SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 3).
Iteration: 1 / 12000 [ 0%] (Warmup)
Iteration: 1200 / 12000 [ 10%] (Warmup)
Iteration: 2400 / 12000 [ 20%] (Sampling)
Iteration: 3600 / 12000 [ 30%] (Sampling)
Iteration: 4800 / 12000 [ 40%] (Sampling)
Iteration: 6000 / 12000 [ 50%] (Sampling)
Iteration: 7200 / 12000 [ 60%] (Sampling)
Iteration: 8400 / 12000 [ 70%] (Sampling)
Iteration: 9600 / 12000 [ 80%] (Sampling)
Iteration: 10800 / 12000 [ 90%] (Sampling)
Iteration: 12000 / 12000 [100%] (Sampling)
Elapsed Time: 0.91 seconds (Warm-up)
2.57 seconds (Sampling)
3.48 seconds (Total)
> print(data.rstan)
Inference for Stan model: rstanString.
3 chains, each with iter=12000; warmup=2000; thin=10;
post-warmup draws per chain=1000, total post-warmup draws=3000.
mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat
beta[1] 42.3 0.1 7.2 28.2 37.7 42.4 46.8 56.7 3000 1
beta[2] 29.8 0.2 10.0 9.2 23.6 29.6 36.3 49.3 3000 1
beta[3] 37.0 0.2 10.0 16.8 31.0 37.0 43.4 56.7 3000 1
sigma 4.4 0.0 0.3 3.9 4.2 4.4 4.6 5.0 3000 1
sigma_S 15.3 0.1 3.7 10.0 12.6 14.6 17.2 24.1 3000 1
lp__ -340.3 0.1 3.3 -347.6 -342.4 -340.0 -337.9 -334.8 2994 1
Samples were drawn using NUTS(diag_e) at Fri Mar 14 12:13:25 2014.
For each parameter, n_eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor on split chains (at
convergence, Rhat=1).
> > > > data.rstan.df <-as.data.frame(extract(data.rstan)) > head(data.rstan.df)
beta.1 beta.2 beta.3 sigma sigma_S lp__ 1 39.30 21.756 36.29 4.654 22.44 -342.1 2 40.49 29.395 36.92 4.509 11.68 -342.4 3 63.87 2.698 24.49 4.545 19.39 -342.1 4 34.65 43.400 42.36 4.359 14.48 -336.7 5 53.57 18.175 20.04 4.577 16.14 -340.4 6 35.60 47.025 38.24 4.370 19.74 -344.5
> > data.mcmc<-rstan:::as.mcmc.list.stanfit(data.rstan) > > > plyr:::adply(as.matrix(data.rstan.df),2,MCMCsum)
X1 Median X0. X25. X50. X75. X100. lower upper lower.1 upper.1 1 beta.1 42.410 13.306 37.67 42.410 46.782 70.19 28.044 56.341 36.125 49.511 2 beta.2 29.618 -13.250 23.64 29.618 36.251 70.28 9.234 49.360 20.053 38.706 3 beta.3 36.967 -1.784 31.00 36.967 43.437 71.64 16.677 56.508 27.561 46.028 4 sigma 4.397 3.591 4.23 4.397 4.581 5.59 3.894 4.937 4.121 4.639 5 sigma_S 14.624 7.465 12.58 14.624 17.229 40.27 9.306 22.635 10.730 17.069 6 lp__ -339.990 -355.651 -342.36 -339.990 -337.936 -332.57 -347.156 -334.518 -342.958 -336.622
> rstanString=" data{ int n; int nA; int nSites; vector [n] y; matrix [nSites,nA] X; matrix [n,nSites] Z; } parameters{ vector[nA] beta; vector[nSites] gamma; real<lower=0> sigma; real<lower=0> sigma_S; } model{ vector [n] mu_site; vector [nSites] mu; // Priors beta ~ normal( 0 , 1000 ); gamma ~ normal( 0 , 1000 ); sigma ~ uniform( 0 , 100 ); sigma_S~ uniform( 0 , 100 ); mu_site <- Z*gamma; y ~ normal( mu_site , sigma ); mu <- X*beta; gamma ~ normal(mu, sigma_S); } generated quantities { vector [n] mu_site; vector [nSites] mu; vector [n] y_err; real sd_y; vector [nSites] mu_site_err; real sd_site; real sd_A; mu_site <- Z*gamma; y_err <- mu_site - y; sd_y <- sd(y_err); mu <- X*beta; mu_site_err <- mu - gamma; sd_site <- sd(mu_site_err); sd_A <- sd(beta); } "
Unlike JAGS, STAN's var() and sd() functions generate sample variance and standard deviation rather than population versions. Therefore, it is arguably better to calculate the population standard deviations within R from the MCMC samples.
> dt.A <- ddply(data,~Sites,catcolwise(unique)) > X<-model.matrix(~A, dt.A) > Z<-model.matrix(~Sites-1, data) > data.list <- list(y=data$y, X=X, Z=Z, n=nrow(data), nSites=nrow(X),nA=ncol(X)) > burnInSteps = 3000 > nChains = 3 > numSavedSteps = 3000 > thinSteps = 10 > nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) > library(rstan) > rstan.SD.time <- system.time( + data.rstanSD <- stan(data=data.list, + model_code=rstanString, + pars=c('beta','sigma','sigma_S','sd_A','sd_site','sd_y'), + chains=nChains, + iter=nIter, + warmup=burnInSteps, + thin=thinSteps, + save_dso=TRUE + ) + )
TRANSLATING MODEL 'rstanString' FROM Stan CODE TO C++ CODE NOW.
COMPILING THE C++ CODE FOR MODEL 'rstanString' NOW.
SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 1).
Iteration: 1 / 13000 [ 0%] (Warmup)
Iteration: 1300 / 13000 [ 10%] (Warmup)
Iteration: 2600 / 13000 [ 20%] (Warmup)
Iteration: 3900 / 13000 [ 30%] (Sampling)
Iteration: 5200 / 13000 [ 40%] (Sampling)
Iteration: 6500 / 13000 [ 50%] (Sampling)
Iteration: 7800 / 13000 [ 60%] (Sampling)
Iteration: 9100 / 13000 [ 70%] (Sampling)
Iteration: 10400 / 13000 [ 80%] (Sampling)
Iteration: 11700 / 13000 [ 90%] (Sampling)
Iteration: 13000 / 13000 [100%] (Sampling)
Elapsed Time: 1.32 seconds (Warm-up)
3.02 seconds (Sampling)
4.34 seconds (Total)
SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 2).
Iteration: 1 / 13000 [ 0%] (Warmup)
Iteration: 1300 / 13000 [ 10%] (Warmup)
Iteration: 2600 / 13000 [ 20%] (Warmup)
Iteration: 3900 / 13000 [ 30%] (Sampling)
Iteration: 5200 / 13000 [ 40%] (Sampling)
Iteration: 6500 / 13000 [ 50%] (Sampling)
Iteration: 7800 / 13000 [ 60%] (Sampling)
Iteration: 9100 / 13000 [ 70%] (Sampling)
Iteration: 10400 / 13000 [ 80%] (Sampling)
Iteration: 11700 / 13000 [ 90%] (Sampling)
Iteration: 13000 / 13000 [100%] (Sampling)
Elapsed Time: 1.01 seconds (Warm-up)
3.11 seconds (Sampling)
4.12 seconds (Total)
SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 3).
Iteration: 1 / 13000 [ 0%] (Warmup)
Iteration: 1300 / 13000 [ 10%] (Warmup)
Iteration: 2600 / 13000 [ 20%] (Warmup)
Iteration: 3900 / 13000 [ 30%] (Sampling)
Iteration: 5200 / 13000 [ 40%] (Sampling)
Iteration: 6500 / 13000 [ 50%] (Sampling)
Iteration: 7800 / 13000 [ 60%] (Sampling)
Iteration: 9100 / 13000 [ 70%] (Sampling)
Iteration: 10400 / 13000 [ 80%] (Sampling)
Iteration: 11700 / 13000 [ 90%] (Sampling)
Iteration: 13000 / 13000 [100%] (Sampling)
Elapsed Time: 1.13 seconds (Warm-up)
2.09 seconds (Sampling)
3.22 seconds (Total)
> print(data.rstanSD)
Inference for Stan model: rstanString.
3 chains, each with iter=13000; warmup=3000; thin=10;
post-warmup draws per chain=1000, total post-warmup draws=3000.
mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat
beta[1] 42.3 0.1 7.2 28.4 37.7 42.4 47.0 56.4 3000 1
beta[2] 29.9 0.2 10.1 10.1 23.7 29.9 36.1 50.4 2776 1
beta[3] 37.0 0.2 10.2 16.6 30.7 37.0 43.4 57.3 2748 1
sigma 4.4 0.0 0.3 3.9 4.2 4.4 4.6 5.0 2964 1
sigma_S 15.4 0.1 3.7 10.1 12.7 14.7 17.2 24.6 3000 1
sd_A 10.3 0.1 6.0 1.6 6.0 9.3 13.5 23.9 2824 1
sd_site 13.9 0.0 1.5 12.3 13.0 13.5 14.4 17.8 2930 1
sd_y 4.4 0.0 0.1 4.2 4.3 4.4 4.4 4.6 2555 1
lp__ -340.4 0.1 3.4 -348.2 -342.6 -340.0 -338.0 -334.7 2791 1
Samples were drawn using NUTS(diag_e) at Fri Mar 14 12:16:09 2014.
For each parameter, n_eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor on split chains (at
convergence, Rhat=1).
> > > > data.rstanSD.df <-as.data.frame(extract(data.rstanSD)) > head(data.rstanSD.df)
beta.1 beta.2 beta.3 sigma sigma_S sd_A sd_site sd_y lp__ 1 45.59 17.86 39.89 4.353 15.58 14.644 14.09 4.323 -337.0 2 54.16 39.96 16.57 4.727 35.32 18.980 17.94 4.375 -347.4 3 37.41 38.08 43.25 5.241 15.74 3.197 13.96 4.437 -344.4 4 49.02 21.05 35.86 4.865 11.55 13.993 13.09 4.364 -339.8 5 51.12 13.01 30.65 4.384 18.00 19.071 14.42 4.403 -340.5 6 49.26 13.92 37.52 4.543 14.01 17.997 14.98 4.564 -346.8
> > data.mcmcSD<-rstan:::as.mcmc.list.stanfit(data.rstanSD) > > > plyr:::adply(as.matrix(data.rstanSD.df),2,MCMCsum)
X1 Median X0. X25. X50. X75. X100. lower upper lower.1 upper.1 1 beta.1 42.400 13.59283 37.673 42.400 47.031 73.862 29.167 56.921 35.342 49.033 2 beta.2 29.934 -30.63715 23.659 29.934 36.146 70.804 9.640 49.212 21.047 39.742 3 beta.3 37.008 -19.74213 30.676 37.008 43.405 74.011 14.478 55.092 28.758 47.760 4 sigma 4.404 3.55402 4.216 4.404 4.588 5.540 3.906 4.947 4.092 4.629 5 sigma_S 14.742 7.16649 12.687 14.742 17.168 38.677 9.372 22.643 10.937 17.182 6 sd_A 9.344 0.09846 5.978 9.344 13.515 52.484 1.137 22.056 3.197 13.639 7 sd_site 13.521 11.58517 12.987 13.521 14.419 33.711 11.901 16.695 12.340 14.202 8 sd_y 4.367 4.19310 4.318 4.367 4.424 4.834 4.224 4.538 4.280 4.433 9 lp__ -340.013 -355.96610 -342.561 -340.013 -337.953 -332.525 -346.853 -333.791 -343.342 -336.789
>
Quick glance Bayesian regression model specification
| JAGS Full | JAGS Matrix | JAGS heirarchy | Stan Cellmeans | Stan Full | Stan Matrix | Stan hierarcical | |
|---|---|---|---|---|---|---|---|
| Iterations | 13000 | 13000 | 13000 | 13000 | 13000 | 13000 | 12000 |
| Thinning | 10 | 10 | 10 | 10 | 10 | 10 | 10 |
| Samples | 3000 | 3000 | 3000 | 3000 | 3000 | 3000 | 3000 |
| Maximum ACF | 0.0098 | 0.0278 | 0.0379 | 0.0669 | 0.0497 | 0.0225 | 0.0282 |
| Intercept | 42.41 [28.69 , 56.54] | 42.45 [28.63 , 56.38] | 42.12 [28.53 , 55.78] | 42.16 [26.69 , 55.20] | 42.10 [28.58 , 56.32] | 42.30 [28.76 , 56.78] | 42.41 [28.04 , 56.34] |
| alpha2 | 29.74 [7.81 , 48.57] | 29.66 [9.97 , 49.27] | 29.96 [10.30 , 48.86] | 71.91 [58.01 , 86.16] | 29.99 [10.43 , 49.77] | 29.68 [9.76 , 49.23] | 29.62 [9.23 , 49.36] |
| alpha3 | 37.00 [15.87 , 56.34] | 37.16 [17.70 , 56.74] | 37.30 [17.29 , 56.74] | 78.86 [65.06 , 92.76] | 37.23 [17.55 , 57.08] | 37.05 [14.79 , 55.64] | 36.97 [16.68 , 56.51] |
| sigma | 4.40 [3.90 , 4.94] | 4.40 [3.89 , 4.94] | 4.39 [3.89 , 4.92] | 4.40 [3.91 , 4.97] | 4.41 [3.94 , 5.00] | 4.40 [3.87 , 4.93] | 4.40 [3.89 , 4.94] |
| sigma.site | 14.73 [9.37 , 22.30] | 14.71 [9.12 , 22.54] | 14.74 [9.33 , 22.70] | 14.62 [9.44 , 22.45] | 14.78 [9.40 , 22.29] | 14.76 [9.40 , 22.74] | 14.62 [9.31 , 22.63] |
| Time (user self) | 3.89 | 4.10 | 3.48 | 15.46 | 50.61 | 85.79 | 10.92 |
| Time (system self) | 0.00 | 0.00 | 0.00 | 0.01 | 0.03 | 0.03 | 0.04 |
| Time (elapsed) | 3.93 | 4.10 | 3.49 | 42.87 | 78.70 | 116.36 | 39.48 |
| Time (user child) | 0.00 | 0.00 | 0.00 | 26.98 | 27.48 | 29.67 | 28.03 |
| Time (system child) | 0.00 | 0.00 | 0.00 | 0.59 | 0.60 | 0.63 | 0.68 |
| Model | JAGS | rstan |
|---|---|---|
| Full effects parameterization |
model {
#Likelihood
for (i in 1:n) {
y[i]~dnorm(mu[i],tau)
mu[i] <- alpha0 + alpha[A[i]] + beta[site[i]]
}
#Priors
alpha0 ~ dnorm(0, 1.0E-6)
alpha[1] <- 0
for (i in 2:nA) {
alpha[i] ~ dnorm(0, 1.0E-6) #prior
}
for (i in 1:nSite) {
beta[i] ~ dnorm(0, tau.B) #prior
}
sigma ~ dunif(0, 100)
tau <- 1 / (sigma * sigma)
sigma.B ~ dunif(0, 100)
tau.B <- 1 / (sigma.B * sigma.B)
}
|
data{
int n;
int nB;
vector [n] y;
int A2[n];
int A3[n];
int B[n];
}
parameters{
real alpha0;
real alpha2;
real alpha3;
real<lower=0> sigma;
vector [nB] beta;
real<lower=0> sigma_B;
}
model{
real mu[n];
// Priors
alpha0 ~ normal( 0 , 100 );
alpha2 ~ normal( 0 , 100 );
alpha3 ~ normal( 0 , 100 );
beta ~ normal( 0 , sigma_B );
sigma_B ~ uniform( 0 , 100 );
sigma ~ uniform( 0 , 100 );
for ( i in 1:n ) {
mu[i] <- alpha0 + alpha2*A2[i] +
alpha3*A3[i] + beta[B[i]];
}
y ~ normal( mu , sigma );
|
| Matrix parameterization |
model {
#Likelihood
for (i in 1:n) {
y[i]~dnorm(mu[i],tau)
mu[i] <- inprod(alpha[],X[i,]) + beta[site[i]]
}
#Priors
alpha ~ dmnorm(a0,A0)
for (i in 1:nSite) {
beta[i] ~ dnorm(0, tau.B) #prior
}
sigma ~ dunif(0, 100)
tau <- 1 / (sigma * sigma)
sigma.B ~ dunif(0, 100)
tau.B <- 1 / (sigma.B * sigma.B)
}
"
writeLines(modelString,con="../downloads/BUGSscripts/tut9.7bS6.1.m.txt")
|
data{
int n;
int nB;
int nA;
vector [n] y;
matrix [n,nA] X;
int B[n];
vector [nA] a0;
matrix [nA,nA] A0;
}
parameters{
vector [nA] alpha;
real<lower=0> sigma;
vector [nB] beta;
real<lower=0> sigma_B;
}
model{
real mu[n];
// Priors
//alpha ~ normal( 0 , 100 );
alpha ~ multi_normal(a0,A0);
beta ~ normal( 0 , sigma_B );
sigma_B ~ uniform( 0 , 100 );
sigma ~ uniform( 0 , 100 );
for ( i in 1:n ) {
mu[i] <- dot_product(X[i],alpha) + beta[B[i]];
}
|
| Hierarchical parameterization |
model {
#Likelihood (esimating site means (gamma.site)
for (i in 1:n) {
y[i]~dnorm(quad.means[i],tau)
quad.means[i] <- gamma.site[site[i]]
}
for (i in 1:s) {
gamma.site[i] ~ dnorm(site.means[i], tau.site)
site.means[i] <- inprod(beta[],A.Xmat[i,])
}
#Priors
for (i in 1:a) {
beta[i] ~ dnorm(0, 1.0E-6) #prior
}
sigma ~ dunif(0, 100)
tau <- 1 / (sigma * sigma)
sigma.site ~ dunif(0, 100)
tau.site <- 1 / (sigma.site * sigma.site)
}
|
data{
int n;
int nA;
int nSites;
vector [n] y;
matrix [nSites,nA] X;
matrix [n,nSites] Z;
}
parameters{
vector[nA] beta;
vector[nSites] gamma;
real<lower=0> sigma;
real<lower=0> sigma_S;
}
model{
vector [n] mu_site;
vector [nSites] mu;
// Priors
beta ~ normal( 0 , 1000 );
gamma ~ normal( 0 , 1000 );
sigma ~ uniform( 0 , 100 );
sigma_S~ uniform( 0 , 100 );
mu_site <- Z*gamma;
y ~ normal( mu_site , sigma );
mu <- X*beta;
gamma ~ normal(mu, sigma_S);
}
|
| Including finite-population standard deviations |
model {
#Likelihood (esimating site means (gamma.site)
for (i in 1:n) {
y[i]~dnorm(quad.means[i],tau)
quad.means[i] <- gamma.site[site[i]]
y.err[i]<- quad.means[i]-y[i]
}
for (i in 1:s) {
gamma.site[i] ~ dnorm(site.means[i], tau.site)
site.means[i] <- inprod(beta[],A.Xmat[i,])
site.err[i] <- site.means[i] - gamma.site[i]
}
#Priors
for (i in 1:a) {
beta[i] ~ dnorm(0, 1.0E-6) #prior
}
sigma ~ dunif(0, 100)
tau <- 1 / (sigma * sigma)
sigma.site ~ dunif(0, 100)
tau.site <- 1 / (sigma.site * sigma.site)
sd.y <- sd(y.err)
sd.site <- sd(site.err)
sd.A <- sd(beta)
}
|
data{
int n;
int nA;
int nSites;
vector [n] y;
matrix [nSites,nA] X;
matrix [n,nSites] Z;
}
parameters{
vector[nA] beta;
vector[nSites] gamma;
real<lower=0> sigma;
real<lower=0> sigma_S;
}
model{
vector [n] mu_site;
vector [nSites] mu;
// Priors
beta ~ normal( 0 , 1000 );
gamma ~ normal( 0 , 1000 );
sigma ~ uniform( 0 , 100 );
sigma_S~ uniform( 0 , 100 );
mu_site <- Z*gamma;
y ~ normal( mu_site , sigma );
mu <- X*beta;
gamma ~ normal(mu, sigma_S);
}
generated quantities {
vector [n] mu_site;
vector [nSites] mu;
vector [n] y_err;
real sd_y;
vector [nSites] mu_site_err;
real sd_site;
real sd_A;
mu_site <- Z*gamma;
y_err <- mu_site - y;
sd_y <- sd(y_err);
mu <- X*beta;
mu_site_err <- mu - gamma;
sd_site <- sd(mu_site_err);
sd_A <- sd(beta);
}
|
Scenario and Data
Now imagine a similar experiment in which we intend to measure a response ($y$) to one of treatments (three levels; 'a1', 'a2' and 'a3'). As with the previous design, we decided to establish a nested design in which there are sub-replicate (1m Quadrats) within each Site.
In the current design, we have decided to further sub-replicate. Within each of the 5 Quadrats, we are going to randomly place 2x10cm pit traps. Now we have Sites nested within Treatments, Quadrats nested within Sites AND, Pits nested within Sites. The latter of these (Pits nested within Sites) are the observations ($y$). As this section is mainly about the generation of artificial data (and not specifically about what to do with the data), understanding the actual details are optional and can be safely skipped. Consequently, I have folded (toggled) this section away.
- the number of treatments = 3
- the number of sites per treatment = 5
- the total number of sites = 15
- the number of quadrats per site = 5
- the number of pits per quadrat = 2
- the mean of the treatments = 40, 70 and 80 respectively
- the variability (standard deviation) between sites of the same treatment = 15
- the variability (standard deviation) between quadrats within sites = 10
- the variability (standard deviation) between pits within quadrats = 5
> set.seed(3) > nTreat <- 3 > nSites <- 15 > nSitesPerTreat <- nSites/nTreat > nQuads <- 5 > nPits <- 2 > site.sigma <- 10 # sd within between sites within treatment > quad.sigma <- 10 > sigma <- 7.5 > n <- nSites * nQuads * nPits > sites <- gl(n = nSites, n/nSites, n, lab = paste("site", 1:nSites)) > A <- gl(nTreat, n/nTreat, n, labels = c("a1", "a2", "a3")) > a.means <- c(40, 70, 80) > > # A<-gl(nTreat,nSites/nTreat,nSites,labels=c('a1','a2','a3')) > a <- gl(nTreat, 1, nTreat, labels = c("a1", "a2", "a3")) > a.X <- model.matrix(~a, expand.grid(a)) > a.eff <- as.vector(solve(a.X, a.means)) > site.means <- rnorm(nSites, A.X %*% a.eff, site.sigma)
Error: object 'A.X' not found
> > A <- gl(nTreat, nSites/nTreat, nSites, labels = c("a1", "a2", "a3")) > A.X <- model.matrix(~A, expand.grid(A)) > # a.X <- model.matrix(~A, expand.grid(A=gl(nTreat,nSites/nTreat,nSites,labels=c('a1','a2','a3')))) > site.means <- rnorm(nSites, A.X %*% a.eff, site.sigma) > > SITES <- gl(nSites, (nSites * nQuads)/nSites, nSites * nQuads, labels = paste("site", 1:nSites)) > sites.X <- model.matrix(~SITES - 1) > quad.means <- rnorm(nSites * nQuads, sites.X %*% site.means, quad.sigma) > > # SITES <- gl(nSites,1,nSites,labels=paste('site',1:nSites)) sites.X <- model.matrix(~SITES-1) quad.means <- rnorm(nSites*nQuads,sites.X %*% site.means,quad.sigma) > > QUADS <- gl(nSites * nQuads, n/(nSites * nQuads), n, labels = paste("quad", 1:(nSites * nQuads))) > quads.X <- model.matrix(~QUADS - 1) > # quads.eff <- as.vector(solve(quads.X,quad.means)) pit.means <- rnorm(n,quads.eff %*% t(quads.X),sigma) > pit.means <- rnorm(n, quads.X %*% quad.means, sigma) > > PITS <- gl(nPits * nSites * nQuads, 1, n, labels = paste("pit", 1:(nPits * nSites * nQuads))) > data1 <- data.frame(Pits = PITS, Quads = QUADS, Sites = rep(SITES, each = 2), A = rep(A, each = nQuads * nPits), y = pit.means) > # data1<-data1[order(data1$A,data1$Sites,data1$Quads),] > head(data1) #print out the first six rows of the data set
Pits Quads Sites A y 1 pit 1 quad 1 site 1 a1 20.90 2 pit 2 quad 1 site 1 a1 19.88 3 pit 3 quad 2 site 1 a1 15.97 4 pit 4 quad 2 site 1 a1 28.76 5 pit 5 quad 3 site 1 a1 20.97 6 pit 6 quad 3 site 1 a1 23.37
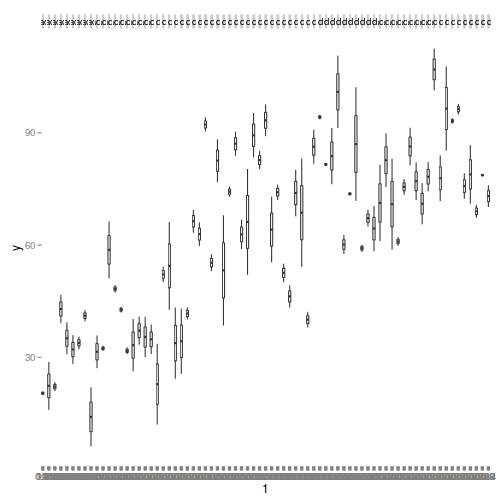
> library(ggplot2) > ggplot(data1, aes(y = y, x = 1)) + geom_boxplot() + facet_grid(. ~ Quads)
> > comparisonTab <- NULL
Exploratory data analysis
Normality and Homogeneity of variance
> # Effects of treatment > library(plyr) > boxplot(y ~ A, ddply(data1, ~A + Sites, numcolwise(mean, na.rm = T)))
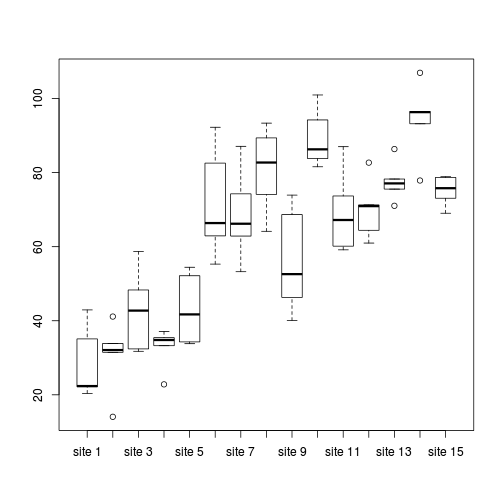
> > # Site effects > boxplot(y ~ Sites, ddply(data1, ~A + Sites + Quads, numcolwise(mean, na.rm = T)))
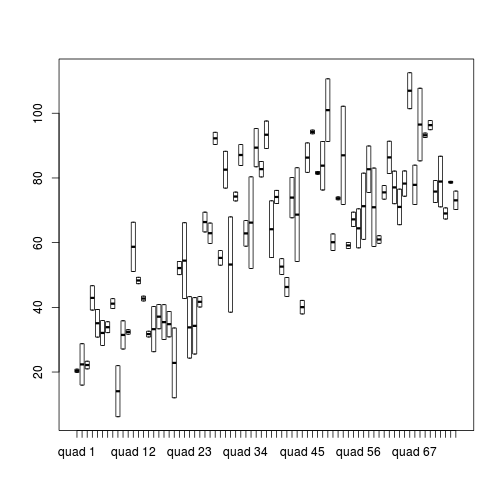
> > # Quadrat effects > boxplot(y ~ Quads, ddply(data1, ~A + Sites + Quads + Pits, numcolwise(mean, na.rm = T)))
Conclusions:
- there is no evidence that the response variable is consistently non-normal across all populations - each boxplot is approximately symmetrical
- there is no evidence that variance (as estimated by the height of the boxplots) differs between the five populations. . More importantly, there is no evidence of a relationship between mean and variance - the height of boxplots does not increase with increasing position along the y-axis. Hence it there is no evidence of non-homogeneity
- It is a little difficult to assess normality/homogeneity of variance of quadrats since there are only two pits per quadrat. Nevertheless, there is no suggestion that variance increases with increasing mean.
- transform the scale of the response variables (to address normality etc). Note transformations should be applied to the entire response variable (not just those populations that are skewed).
JAGS
| Full parameterization | Matrix parameterization | Heirarchical parameterization |
|---|---|---|
| $$ \begin{array}{rcl} y_{ijk}&\sim&\mathcal{N}(\mu_{ij},\sigma^2)\\ \mu_{ijk} &=& \alpha_0 + \alpha_{i} + \beta.s_{j(i)} + \beta.q_{k(j(i))}\\ \beta.s_{j(i)}&\sim&\mathcal{N}(0,\sigma_{s}^2)\\ \beta.q_{k(j(i))}&\sim&\mathcal{N}(0,\sigma_{q}^2)\\ \alpha_0, \alpha_i&\sim&\mathcal{N}(0,100000)\\ \sigma^2,\sigma_{s}^2,\sigma_{q}^2&\sim&\mathcal{U}(0,100)\\ \end{array} $$ | $$ \begin{array}{rcl} y_{ijk}&\sim&\mathcal{N}(\mu_{ij},\sigma^2)\\ \mu_{ij} &=& \alpha\mathbf{X} + \beta.s_{j(i)} + \beta.q_{k(j(i))}\\ \beta_{j(i)}&\sim&\mathcal{N}(0,\sigma_{s}^2)\\ \beta_{k(j(i)}&\sim&\mathcal{N}(0,\sigma_{q}^2)\\ \alpha&\sim&\mathcal{MVN}(0,100000)\\ \sigma^2,\sigma_{s}^2,\sigma_{q}^2&\sim&\mathcal{U}(0,100)\\ \end{array} $$ | $$ \begin{array}{rcl} y_{ijk}&\sim &\mathcal{N}(\mu_{kji}, \sigma^2)\\ \mu_{kji}&=&\beta.q_{k(j(i))}\\ \beta.q_{k(j(i))}&\sim&\mathcal{N}(\mu.q_{i},\sigma_{q}^2)\\ \mu.q_{j(i)}&=&\beta.s_{j(i)}\\ \beta.s_{j(i)}&\sim &\mathcal{N}(\mu.s_{j(i)}, \sigma_{s}^2)\\ \mu.s_{i}&=&\alpha\mathbf{X}\\ \alpha_i&\sim&\mathcal{N}(0, 1000000)\\ \sigma^2&\sim&\mathcal{U}(0,100)\\ \sigma_{B}^2&\sim&\mathcal{U}(0,100) \end{array} $$ |
Full effects parameterization
> modelString=" model { #Likelihood for (i in 1:n) { y[i]~dnorm(mu[i],tau) mu[i] <- alpha0 + alpha[A[i]] + beta.s[site[i]] + beta.q[quad[i]] } #Priors alpha0 ~ dnorm(0, 1.0E-6) alpha[1] <- 0 for (i in 2:nA) { alpha[i] ~ dnorm(0, 1.0E-6) #prior } for (i in 1:nSite) { beta.s[i] ~ dnorm(0, tau.site) #prior } for (i in 1:nQuad) { beta.q[i] ~ dnorm(0, tau.quad) #prior } sigma ~ dunif(0, 100) tau <- 1 / (sigma * sigma) sigma.site ~ dunif(0, 100) tau.site <- 1 / (sigma.site * sigma.site) sigma.quad ~ dunif(0, 100) tau.quad <- 1 / (sigma.quad * sigma.quad) } " > writeLines(modelString,con="../downloads/BUGSscripts/tut9.7bT6.1.f.txt")
> data1.list <- with(data1, + list(y=y, + site=as.numeric(Sites), + quad=as.numeric(Quads), + A=as.numeric(A), + n=nrow(data1), + nSite=length(levels(Sites)), + nQuad=length(levels(Quads)), + nA = length(levels(A)) + ) + ) > > params <- c("alpha0","alpha","sigma","sigma.site","sigma.quad") > adaptSteps = 1000 > burnInSteps = 3000 > nChains = 3 > numSavedSteps = 3000 > thinSteps = 10 > nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) > > library(R2jags) > rnorm(1)
[1] 0.8568
> jags.effects.f.time <- system.time( + data.r2jags.f <- jags(data=data1.list, + inits=NULL, + parameters.to.save=params, + model.file="../downloads/BUGSscripts/tut9.7bT6.1.f.txt", + n.chains=3, + n.iter=nIter, + n.burnin=burnInSteps, + n.thin=thinSteps + ) + )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 786 Initializing model
> jags.effects.f.time
user system elapsed 7.508 0.000 7.546
> print(data.r2jags.f)
Inference for Bugs model at "../downloads/BUGSscripts/tut9.7bT6.1.f.txt", fit using jags,
3 chains, each with 13000 iterations (first 3000 discarded), n.thin = 10
n.sims = 3000 iterations saved
mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff
alpha[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.00 1.000 1
alpha[2] 37.733 7.319 23.497 33.126 37.675 42.257 53.15 1.002 1600
alpha[3] 41.568 7.359 27.223 36.886 41.432 46.113 56.18 1.002 1300
alpha0 35.646 5.179 25.601 32.501 35.716 38.956 45.88 1.004 580
sigma 8.853 0.734 7.580 8.326 8.791 9.307 10.46 1.001 3000
sigma.quad 8.425 1.325 5.850 7.554 8.381 9.289 11.08 1.002 1200
sigma.site 10.130 3.007 5.388 8.037 9.681 11.835 16.74 1.002 1700
deviance 1078.239 18.108 1045.628 1065.457 1077.185 1089.555 1116.62 1.001 2000
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 163.9 and DIC = 1242.1
DIC is an estimate of expected predictive error (lower deviance is better).
> data.mcmc.list.f <- as.mcmc(data.r2jags.f) > Data.mcmc.list.f <- data.mcmc.list.f
[1] "alpha[1]" "alpha[2]" "alpha[3]" "alpha0" "deviance" "sigma" "sigma.quad" "sigma.site"
Error: obj must have nsamp > 1
Matrix parameterization
> modelString=" model { #Likelihood for (i in 1:n) { y[i]~dnorm(mu[i],tau) mu[i] <- inprod(alpha[],X[i,]) + beta.site[site[i]] + beta.quad[quad[i]] } #Priors alpha ~ dmnorm(a0,A0) for (i in 1:nSite) { beta.site[i] ~ dnorm(0, tau.site) #prior } for (i in 1:nQuad) { beta.quad[i] ~ dnorm(0, tau.quad) #prior } sigma ~ dunif(0, 100) tau <- 1 / (sigma * sigma) sigma.site ~ dunif(0, 100) tau.site <- 1 / (sigma.site * sigma.site) sigma.quad ~ dunif(0, 100) tau.quad <- 1 / (sigma.quad * sigma.quad) } " > writeLines(modelString,con="../downloads/BUGSscripts/tut9.7bT6.1.m.txt")
> A.Xmat <- model.matrix(~A,data) > nA <- ncol(A.Xmat) > data1.list <- with(data1, + list(y=y, + site=as.numeric(Sites), + quad=as.numeric(Quads), + X=A.Xmat, + n=nrow(data1), + nSite=length(levels(Sites)), + nQuad=length(levels(Quads)), + a0=rep(0,nA), A0=diag(1.0E-06,nA) + ) + ) > > params <- c("alpha","sigma","sigma.site", "sigma.quad") > adaptSteps = 1000 > burnInSteps = 3000 > nChains = 3 > numSavedSteps = 3000 > thinSteps = 10 > nIter = ceiling((numSavedSteps * thinSteps)/nChains) > > library(R2jags) > rnorm(1)
[1] 1.141
> jags1.effects.m.time <- system.time( + data1.r2jags.m <- jags(data=data1.list, + inits=NULL, + parameters.to.save=params, + model.file="../downloads/BUGSscripts/tut9.7bT6.1.m.txt", + n.chains=3, + n.iter=nIter, + n.burnin=burnInSteps, + n.thin=thinSteps + ) + )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 1248 Initializing model
> jags.effects.m.time
user system elapsed 65.168 0.136 66.346
> print(data1.r2jags.m)
Inference for Bugs model at "../downloads/BUGSscripts/tut9.7bT6.1.m.txt", fit using jags,
3 chains, each with 10000 iterations (first 3000 discarded), n.thin = 10
n.sims = 2100 iterations saved
mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff
alpha[1] 35.465 5.157 25.502 32.189 35.463 38.647 46.19 1.002 1300
alpha[2] 37.971 7.192 24.208 33.242 37.791 42.611 52.31 1.001 2100
alpha[3] 41.883 7.254 27.861 37.230 41.737 46.391 56.59 1.001 2100
sigma 8.882 0.741 7.552 8.365 8.839 9.376 10.50 1.002 1100
sigma.quad 8.474 1.339 6.029 7.542 8.401 9.352 11.19 1.002 1900
sigma.site 9.992 2.884 5.598 7.961 9.622 11.475 16.79 1.000 2100
deviance 1078.104 17.534 1046.092 1065.723 1077.542 1090.099 1114.26 1.003 860
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 153.5 and DIC = 1231.6
DIC is an estimate of expected predictive error (lower deviance is better).
> data1.mcmc.list.m <- as.mcmc(data1.r2jags.m) > Data1.mcmc.list.m <- data1.mcmc.list.m
Hierarchical parameterization
> modelString=" model { #Likelihood for (i in 1:n) { y[i]~dnorm(pit.means[i],tau) pit.means[i] <- beta.q[quad[i]] } for (j in 1:q) { beta.q[j] ~ dnorm(quad.means[j], tau.quad) quad.means[j] <- beta.s[site[j]] } for (k in 1:s) { beta.s[k] ~ dnorm(site.means[k], tau.site) site.means[k] <- inprod(alpha[],A.Xmat[k,]) } ##Priors and derivatives alpha ~ dmnorm(a0,A0) sigma ~ dunif(0,100) tau <- 1 / (sigma * sigma) sigma.quad ~ dunif(0,100) tau.quad <- 1 / (sigma.quad * sigma.quad) sigma.site ~ dunif(0,100) tau.site <- 1 / (sigma.site * sigma.site) } " > writeLines(modelString,con="../downloads/BUGSscripts/tut9.7bS6.2.txt") > > A.Xmat <- model.matrix(~A,ddply(data1, ~Sites, function(x) data.frame(A=unique(x$A)))) > S <- ddply(data1, ~Sites*Quads, function(x) data.frame(Sites=unique(x$Sites)))$Sites > #Quadrats must have unique names within each site! > Q <- ddply(data1, ~Sites*Quads*Pits, function(x) data.frame(Quads=unique(x$Quads)))$Quads > nA <- ncol(A.Xmat) > data1.list <- with(data1, + list(y=y, + site=as.numeric(S), + quad=as.numeric(Q), + A.Xmat= A.Xmat, + n=nrow(data1), + s=length(unique(S)), + q=length(unique(Q)), + a0=rep(0,nA), + A0=diag(1.0E-06,nA) + ) + ) > > params <- c("alpha","sigma","sigma.site","sigma.quad") > adaptSteps = 1000 > burnInSteps = 3000 > nChains = 3 > numSavedSteps = 3000 > thinSteps = 10 > nIter = ceiling((numSavedSteps * thinSteps)/nChains) > > library(R2jags) > rnorm(1)
[1] -0.1739
> jags.1.time <- system.time( + data1.r2jags <- jags(data=data1.list, + inits=NULL, + parameters.to.save=params, + model.file="../downloads/BUGSscripts/tut9.7bS6.2.txt", + n.chains=3, + n.iter=nIter, + n.burnin=burnInSteps, + n.thin=thinSteps + ) + )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 558 Initializing model
> print(data1.r2jags)
Inference for Bugs model at "../downloads/BUGSscripts/tut9.7bS6.2.txt", fit using jags,
3 chains, each with 10000 iterations (first 3000 discarded), n.thin = 10
n.sims = 2100 iterations saved
mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff
alpha[1] 35.692 5.309 25.104 32.386 35.660 38.839 46.77 1.005 1700
alpha[2] 37.644 7.336 22.967 33.170 37.741 42.200 52.10 1.003 1400
alpha[3] 41.511 7.299 27.212 36.962 41.476 46.038 56.16 1.006 670
sigma 8.848 0.749 7.577 8.327 8.786 9.290 10.59 1.001 2100
sigma.quad 8.440 1.333 5.835 7.588 8.400 9.316 11.13 1.002 1900
sigma.site 10.146 3.048 5.666 8.124 9.773 11.584 17.45 1.003 1200
deviance 1077.700 17.857 1046.641 1065.024 1076.686 1088.543 1116.46 1.001 2100
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 159.5 and DIC = 1237.2
DIC is an estimate of expected predictive error (lower deviance is better).
> > data.mcmc.list1 <- as.mcmc(data1.r2jags) > Data.mcmc.list1 <- data.mcmc.list1 > comparisonTab <- cbind(comparisonTab,'JAGS Hierarchical'=comTab(data.mcmc.list1,cols="alpha|sigma",lag=10,jags.1.time)) > comparisonTab
JAGS Matrix JAGS Hierarchical [1,] "10000" "10000" [2,] "10" "10" [3,] "2100" "2100" [4,] "0.1546" "0.1482" [5,] "35.46 [25.92 , 46.62]" "35.66 [24.14 , 45.58]" [6,] "37.79 [24.66 , 52.59]" "37.74 [22.82 , 51.78]" [7,] "41.74 [27.90 , 56.62]" "41.48 [26.96 , 55.73]" [8,] "8.84 [7.41 , 10.27]" "8.79 [7.45 , 10.35]" [9,] "8.40 [5.89 , 10.99]" "8.40 [5.87 , 11.14]" [10,] "9.62 [5.01 , 15.62]" "9.77 [4.99 , 16.02]" [11,] "5.17" "4.14" [12,] "0.01" "0.00" [13,] "5.19" "4.14" [14,] "0.00" "0.00" [15,] "0.00" "0.00"
> modelString= > writeLines(modelString,con="../downloads/BUGSscripts/tut9.7bS6.2SD.txt") > > A.Xmat <- model.matrix(~A,ddply(data1, ~Sites, function(x) data.frame(A=unique(x$A)))) > S <- ddply(data1, ~Sites*Quads, function(x) data.frame(Sites=unique(x$Sites)))$Sites > #Quadrats must have unique names within each site! > Q <- ddply(data1, ~Sites*Quads*Pits, function(x) data.frame(Quads=unique(x$Quads)))$Quads > data1.list <- with(data1, + list(y=y, + site=as.numeric(S), + quad=as.numeric(Q), + A.Xmat= A.Xmat, + n=nrow(data1), + s=length(unique(S)), + q=length(unique(Q)), + a = ncol(A.Xmat) + ) + ) > > params <- c("beta","sigma","sd.y",'sd.quad','sd.site','sd.A') > adaptSteps = 1000 > burnInSteps = 2000 > nChains = 3 > numSavedSteps = 50000 > thinSteps = 10 > nIter = ceiling((numSavedSteps * thinSteps)/nChains) > > library(R2jags) > rnorm(1)
[1] 0.8568
> jags.1SD.time <- system.time( + data.1SD.r2jags <- jags(data=data1.list, + inits=NULL, + parameters.to.save=params, + model.file="../downloads/BUGSscripts/tut9.7bS6.2SD.txt", + n.chains=3, + n.iter=nIter, + n.burnin=burnInSteps, + n.thin=thinSteps + ) + )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 796 Initializing model
> print(data.1SD.r2jags)
Inference for Bugs model at "../downloads/BUGSscripts/tut9.7bS6.2SD.txt", fit using jags,
3 chains, each with 166667 iterations (first 2000 discarded), n.thin = 10
n.sims = 49401 iterations saved
mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff
beta[1] 35.611 5.159 25.392 32.353 35.634 38.825 46.018 1.001 7100
beta[2] 37.795 7.250 23.371 33.234 37.814 42.399 52.105 1.001 12000
beta[3] 41.645 7.277 27.090 37.064 41.668 46.262 56.124 1.001 49000
sd.A 5.537 3.296 0.917 3.131 4.964 7.278 13.503 1.001 49000
sd.quad 8.273 1.094 6.022 7.579 8.307 9.002 10.348 1.001 39000
sd.site 8.800 1.734 5.609 7.684 8.728 9.822 12.479 1.001 49000
sd.y 8.759 0.533 7.850 8.380 8.712 9.084 9.934 1.001 32000
sigma 8.858 0.749 7.541 8.331 8.801 9.327 10.465 1.001 15000
deviance 1078.155 18.181 1045.588 1065.434 1077.045 1089.685 1116.700 1.001 34000
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 165.3 and DIC = 1243.4
DIC is an estimate of expected predictive error (lower deviance is better).
Rstan
I have tried to run an model analogous to that used in the JAGS example above (where each level of the heirarchy informs the next, however it just does not seem to suit STAN. Moreover, matrix multiplication seems to slow sampling down dramatically.
> rstanString=
> X <-model.matrix(~A, data1) > > data1.list <- list(y=data1$y, + Treatments=as.numeric(data1$A), + Sites=as.numeric(data1$Sites), + Quads=as.numeric(data1$Q), + nTreat=3, + nSites=15, + nQuads=75, + n=nrow(data1) + ) > > burnInSteps = 3000 > nChains = 3 > numSavedSteps = 3000 > thinSteps = 10 > nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) > > rstan.1.time <- system.time( + data1.rstan <- stan(data=data1.list, + model_code=rstanString, + pars=c('beta','sigma','sigma_Sites','sigma_Quads'), + chains=nChains, + iter=nIter, + warmup=burnInSteps, + thin=thinSteps, + save_dso=TRUE + ) + )
TRANSLATING MODEL 'rstanString' FROM Stan CODE TO C++ CODE NOW.
COMPILING THE C++ CODE FOR MODEL 'rstanString' NOW.
SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 1).
Iteration: 1 / 13000 [ 0%] (Warmup)
Iteration: 1300 / 13000 [ 10%] (Warmup)
Iteration: 2600 / 13000 [ 20%] (Warmup)
Iteration: 3900 / 13000 [ 30%] (Sampling)
Iteration: 5200 / 13000 [ 40%] (Sampling)
Iteration: 6500 / 13000 [ 50%] (Sampling)
Iteration: 7800 / 13000 [ 60%] (Sampling)
Iteration: 9100 / 13000 [ 70%] (Sampling)
Iteration: 10400 / 13000 [ 80%] (Sampling)
Iteration: 11700 / 13000 [ 90%] (Sampling)
Iteration: 13000 / 13000 [100%] (Sampling)
Elapsed Time: 1.28 seconds (Warm-up)
4.02 seconds (Sampling)
5.3 seconds (Total)
SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 2).
Iteration: 1 / 13000 [ 0%] (Warmup)
Iteration: 1300 / 13000 [ 10%] (Warmup)
Iteration: 2600 / 13000 [ 20%] (Warmup)
Iteration: 3900 / 13000 [ 30%] (Sampling)
Iteration: 5200 / 13000 [ 40%] (Sampling)
Iteration: 6500 / 13000 [ 50%] (Sampling)
Iteration: 7800 / 13000 [ 60%] (Sampling)
Iteration: 9100 / 13000 [ 70%] (Sampling)
Iteration: 10400 / 13000 [ 80%] (Sampling)
Iteration: 11700 / 13000 [ 90%] (Sampling)
Iteration: 13000 / 13000 [100%] (Sampling)
Elapsed Time: 1.45 seconds (Warm-up)
4.07 seconds (Sampling)
5.52 seconds (Total)
SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 3).
Iteration: 1 / 13000 [ 0%] (Warmup)
Iteration: 1300 / 13000 [ 10%] (Warmup)
Iteration: 2600 / 13000 [ 20%] (Warmup)
Iteration: 3900 / 13000 [ 30%] (Sampling)
Iteration: 5200 / 13000 [ 40%] (Sampling)
Iteration: 6500 / 13000 [ 50%] (Sampling)
Iteration: 7800 / 13000 [ 60%] (Sampling)
Iteration: 9100 / 13000 [ 70%] (Sampling)
Iteration: 10400 / 13000 [ 80%] (Sampling)
Iteration: 11700 / 13000 [ 90%] (Sampling)
Iteration: 13000 / 13000 [100%] (Sampling)
Elapsed Time: 1.32 seconds (Warm-up)
4 seconds (Sampling)
5.32 seconds (Total)
> print(data1.rstan)
Inference for Stan model: rstanString.
3 chains, each with iter=13000; warmup=3000; thin=10;
post-warmup draws per chain=1000, total post-warmup draws=3000.
mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat
beta[1] 35.3 0.1 5.2 25.1 32.0 35.4 38.6 45.4 2794 1
beta[2] 73.5 0.1 5.0 63.5 70.3 73.5 76.8 83.5 3000 1
beta[3] 77.3 0.1 5.2 67.0 74.1 77.3 80.5 87.9 3000 1
sigma 8.9 0.0 0.8 7.5 8.3 8.8 9.3 10.5 2771 1
sigma_Sites 10.1 0.1 3.0 5.5 8.0 9.7 11.7 16.9 2985 1
sigma_Quads 8.5 0.0 1.3 5.9 7.6 8.5 9.4 11.2 2702 1
lp__ -633.1 0.2 9.0 -651.8 -639.0 -632.9 -626.7 -616.5 2754 1
Samples were drawn using NUTS(diag_e) at Fri Mar 14 16:38:30 2014.
For each parameter, n_eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor on split chains (at
convergence, Rhat=1).
> > > data1.rstan.df <-as.data.frame(extract(data1.rstan)) > head(data1.rstan.df)
beta.1 beta.2 beta.3 sigma sigma_Sites sigma_Quads lp__ 1 36.71 74.14 73.54 8.617 9.871 9.747 -619.4 2 33.63 76.69 71.35 7.927 7.607 7.718 -625.6 3 27.39 79.37 74.07 7.476 11.117 8.848 -627.8 4 28.38 74.60 74.94 9.205 11.882 7.993 -624.9 5 54.71 78.34 74.42 8.000 16.715 7.632 -636.4 6 34.72 75.58 71.94 8.352 7.711 9.032 -626.3
> > data1.mcmc<-rstan:::as.mcmc.list.stanfit(data1.rstan) > > plyr:::adply(as.matrix(data1.rstan.df),2,MCMCsum)
X1 Median X0. X25. X50. X75. X100. lower upper lower.1 upper.1 1 beta.1 35.363 8.685 32.048 35.363 38.568 57.16 24.586 44.82 30.364 40.201 2 beta.2 73.532 49.248 70.344 73.532 76.772 100.48 64.234 84.11 68.417 77.982 3 beta.3 77.287 51.238 74.114 77.287 80.537 97.19 66.371 87.25 72.068 81.678 4 sigma 8.806 6.819 8.349 8.806 9.338 11.97 7.486 10.40 8.033 9.480 5 sigma_Sites 9.686 2.862 7.997 9.686 11.682 31.49 5.345 16.52 6.340 11.625 6 sigma_Quads 8.467 4.241 7.568 8.467 9.368 13.17 5.777 10.98 7.098 9.737 7 lp__ -632.889 -663.818 -638.966 -632.889 -626.653 -605.88 -649.748 -614.91 -641.153 -623.548
> comparisonTab <- cbind(comparisonTab,'Stan cellmeans'=comTab(data1.mcmc,cols="beta|sigma",lag=10,rstan.1.time)) > comparisonTab
JAGS Matrix JAGS Hierarchical Stan cellmeans [1,] "10000" "10000" "13000" [2,] "10" "10" "10" [3,] "2100" "2100" "3000" [4,] "0.1546" "0.1482" "0.0576" [5,] "35.46 [25.92 , 46.62]" "35.66 [24.14 , 45.58]" "35.36 [24.59 , 44.82]" [6,] "37.79 [24.66 , 52.59]" "37.74 [22.82 , 51.78]" "73.53 [64.23 , 84.11]" [7,] "41.74 [27.90 , 56.62]" "41.48 [26.96 , 55.73]" "77.29 [66.37 , 87.25]" [8,] "8.84 [7.41 , 10.27]" "8.79 [7.45 , 10.35]" "8.81 [7.49 , 10.40]" [9,] "8.40 [5.89 , 10.99]" "8.40 [5.87 , 11.14]" "9.69 [5.35 , 16.52]" [10,] "9.62 [5.01 , 15.62]" "9.77 [4.99 , 16.02]" "8.47 [5.78 , 10.98]" [11,] "5.17" "4.14" "16.35" [12,] "0.01" "0.00" "0.03" [13,] "5.19" "4.14" "2583.75" [14,] "0.00" "0.00" "28.27" [15,] "0.00" "0.00" "0.64"
> rstanString= > X <-model.matrix(~A, data1) > > data1.list <- list(y=data1$y, + X=X, + #Treatments=as.numeric(data1$A), + Sites=as.numeric(data1$Sites), + Quads=as.numeric(data1$Q), + nTreat=3, + nSites=15, + nQuads=75, + n=nrow(data1), + a0=rep(0,3), + A0=diag(1.0E06,3) + ) > > burnInSteps = 3000 > nChains = 3 > numSavedSteps = 3000 > thinSteps = 10 > nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) > > rstan.2.time <- system.time( + data2.rstan <- stan(data=data1.list, + model_code=rstanString, + pars=c('beta','sigma', 'sigma_Sites','sigma_Quads'), + chains=nChains, + iter=nIter, + warmup=burnInSteps, + thin=thinSteps, + save_dso=TRUE + ) + )
TRANSLATING MODEL 'rstanString' FROM Stan CODE TO C++ CODE NOW.
COMPILING THE C++ CODE FOR MODEL 'rstanString' NOW.
SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 1).
Iteration: 1 / 13000 [ 0%] (Warmup)
Iteration: 1300 / 13000 [ 10%] (Warmup)
Iteration: 2600 / 13000 [ 20%] (Warmup)
Iteration: 3900 / 13000 [ 30%] (Sampling)
Iteration: 5200 / 13000 [ 40%] (Sampling)
Iteration: 6500 / 13000 [ 50%] (Sampling)
Iteration: 7800 / 13000 [ 60%] (Sampling)
Iteration: 9100 / 13000 [ 70%] (Sampling)
Iteration: 10400 / 13000 [ 80%] (Sampling)
Iteration: 11700 / 13000 [ 90%] (Sampling)
Iteration: 13000 / 13000 [100%] (Sampling)
Elapsed Time: 5.51 seconds (Warm-up)
11.44 seconds (Sampling)
16.95 seconds (Total)
SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 2).
Iteration: 1 / 13000 [ 0%] (Warmup)
Iteration: 1300 / 13000 [ 10%] (Warmup)
Iteration: 2600 / 13000 [ 20%] (Warmup)
Iteration: 3900 / 13000 [ 30%] (Sampling)
Iteration: 5200 / 13000 [ 40%] (Sampling)
Iteration: 6500 / 13000 [ 50%] (Sampling)
Iteration: 7800 / 13000 [ 60%] (Sampling)
Iteration: 9100 / 13000 [ 70%] (Sampling)
Iteration: 10400 / 13000 [ 80%] (Sampling)
Iteration: 11700 / 13000 [ 90%] (Sampling)
Iteration: 13000 / 13000 [100%] (Sampling)
Elapsed Time: 5.06 seconds (Warm-up)
28.37 seconds (Sampling)
33.43 seconds (Total)
SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 3).
Iteration: 1 / 13000 [ 0%] (Warmup)
Iteration: 1300 / 13000 [ 10%] (Warmup)
Iteration: 2600 / 13000 [ 20%] (Warmup)
Iteration: 3900 / 13000 [ 30%] (Sampling)
Iteration: 5200 / 13000 [ 40%] (Sampling)
Iteration: 6500 / 13000 [ 50%] (Sampling)
Iteration: 7800 / 13000 [ 60%] (Sampling)
Iteration: 9100 / 13000 [ 70%] (Sampling)
Iteration: 10400 / 13000 [ 80%] (Sampling)
Iteration: 11700 / 13000 [ 90%] (Sampling)
Iteration: 13000 / 13000 [100%] (Sampling)
Elapsed Time: 4.69 seconds (Warm-up)
13.59 seconds (Sampling)
18.28 seconds (Total)
> print(data2.rstan)
Inference for Stan model: rstanString.
3 chains, each with iter=13000; warmup=3000; thin=10;
post-warmup draws per chain=1000, total post-warmup draws=3000.
mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat
beta[1] 35.5 0.1 5.2 25.2 32.3 35.5 38.7 45.7 3000 1
beta[2] 37.9 0.1 7.2 23.5 33.3 37.7 42.5 52.6 3000 1
beta[3] 41.8 0.1 7.4 26.5 37.1 41.9 46.4 56.0 3000 1
sigma 8.9 0.0 0.8 7.5 8.3 8.8 9.3 10.5 2974 1
sigma_Sites 10.0 0.1 3.0 5.4 8.0 9.6 11.6 16.7 2634 1
sigma_Quads 8.5 0.0 1.3 6.0 7.6 8.5 9.3 11.1 2717 1
lp__ -631.7 0.2 9.2 -650.5 -637.7 -631.5 -625.2 -615.0 2505 1
Samples were drawn using NUTS(diag_e) at Fri Mar 14 17:48:03 2014.
For each parameter, n_eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor on split chains (at
convergence, Rhat=1).
> > rstan.2.time
user system elapsed 99.121 0.636 99.639
> data1.rstan.m.df <-as.data.frame(extract(data2.rstan)) > > data1.m.mcmc<-rstan:::as.mcmc.list.stanfit(data2.rstan) > > plyr:::adply(as.matrix(data1.rstan.m.df),2,MCMCsum)
X1 Median X0. X25. X50. X75. X100. lower upper lower.1 upper.1 1 beta.1 35.463 14.123 32.303 35.463 38.692 61.34 25.057 45.48 30.700 40.326 2 beta.2 37.696 10.795 33.306 37.696 42.454 68.73 23.821 52.90 30.869 44.211 3 beta.3 41.852 7.796 37.141 41.852 46.377 70.87 27.111 56.19 34.855 48.660 4 sigma 8.819 6.766 8.331 8.819 9.328 11.92 7.480 10.40 7.995 9.449 5 sigma_Sites 9.582 1.955 7.975 9.582 11.560 38.07 5.001 15.85 6.563 11.730 6 sigma_Quads 8.458 2.494 7.574 8.458 9.289 13.44 5.943 11.09 7.127 9.668 7 lp__ -631.452 -668.243 -637.702 -631.452 -625.202 -577.59 -650.261 -614.78 -640.627 -622.636
> comparisonTab <- cbind(comparisonTab,'Stan Matrix'=comTab(data1.m.mcmc,cols="beta|sigma",lag=10,rstan.2.time)) > comparisonTab
JAGS Matrix JAGS Hierarchical Stan cellmeans Stan Matrix [1,] "10000" "10000" "13000" "13000" [2,] "10" "10" "10" "10" [3,] "2100" "2100" "3000" "3000" [4,] "0.1546" "0.1482" "0.0576" "0.0426" [5,] "35.46 [25.92 , 46.62]" "35.66 [24.14 , 45.58]" "35.36 [24.59 , 44.82]" "35.46 [25.06 , 45.48]" [6,] "37.79 [24.66 , 52.59]" "37.74 [22.82 , 51.78]" "73.53 [64.23 , 84.11]" "37.70 [23.82 , 52.90]" [7,] "41.74 [27.90 , 56.62]" "41.48 [26.96 , 55.73]" "77.29 [66.37 , 87.25]" "41.85 [27.11 , 56.19]" [8,] "8.84 [7.41 , 10.27]" "8.79 [7.45 , 10.35]" "8.81 [7.49 , 10.40]" "8.82 [7.48 , 10.40]" [9,] "8.40 [5.89 , 10.99]" "8.40 [5.87 , 11.14]" "9.69 [5.35 , 16.52]" "9.58 [5.00 , 15.85]" [10,] "9.62 [5.01 , 15.62]" "9.77 [4.99 , 16.02]" "8.47 [5.78 , 10.98]" "8.46 [5.94 , 11.09]" [11,] "5.17" "4.14" "16.35" "68.82" [12,] "0.01" "0.00" "0.03" "0.10" [13,] "5.19" "4.14" "2583.75" "99.64" [14,] "0.00" "0.00" "28.27" "30.30" [15,] "0.00" "0.00" "0.64" "0.53"
> rstanString= > X <-model.matrix(~A, data1) > > data1.list <- list(y=data1$y, + Treatments=as.numeric(data1$A), + Sites=as.numeric(data1$Sites), + Quads=as.numeric(data1$Q), + nTreat=3, + nSites=15, + nQuads=75, + n=nrow(data1) + ) > rstan.1.time <- system.time( + data1.rstan <- stan(data=data1.list, + model_code=rstanString, + pars=c('sd_A','sd_Sites','sd_Quads','beta','sigma','sigma_Sites','sigma_Quads'), + chains=nChains, + iter=nIter, + warmup=burnInSteps, + thin=thinSteps, + save_dso=TRUE + ) + )
TRANSLATING MODEL 'rstanString' FROM Stan CODE TO C++ CODE NOW.
COMPILING THE C++ CODE FOR MODEL 'rstanString' NOW.
SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 1).
Iteration: 1 / 166667 [ 0%] (Warmup)
Iteration: 16666 / 166667 [ 9%] (Sampling)
Iteration: 33332 / 166667 [ 19%] (Sampling)
Iteration: 49998 / 166667 [ 29%] (Sampling)
Iteration: 66664 / 166667 [ 39%] (Sampling)
Iteration: 83330 / 166667 [ 49%] (Sampling)
Iteration: 99996 / 166667 [ 59%] (Sampling)
Iteration: 116662 / 166667 [ 69%] (Sampling)
Iteration: 133328 / 166667 [ 79%] (Sampling)
Iteration: 149994 / 166667 [ 89%] (Sampling)
Iteration: 166660 / 166667 [ 99%] (Sampling)
Iteration: 166667 / 166667 [100%] (Sampling)
Elapsed Time: 0.98 seconds (Warm-up)
53.97 seconds (Sampling)
54.95 seconds (Total)
SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 2).
Iteration: 1 / 166667 [ 0%] (Warmup)
Iteration: 16666 / 166667 [ 9%] (Sampling)
Iteration: 33332 / 166667 [ 19%] (Sampling)
Iteration: 49998 / 166667 [ 29%] (Sampling)
Iteration: 66664 / 166667 [ 39%] (Sampling)
Iteration: 83330 / 166667 [ 49%] (Sampling)
Iteration: 99996 / 166667 [ 59%] (Sampling)
Iteration: 116662 / 166667 [ 69%] (Sampling)
Iteration: 133328 / 166667 [ 79%] (Sampling)
Iteration: 149994 / 166667 [ 89%] (Sampling)
Iteration: 166660 / 166667 [ 99%] (Sampling)
Iteration: 166667 / 166667 [100%] (Sampling)
Elapsed Time: 1 seconds (Warm-up)
66.25 seconds (Sampling)
67.25 seconds (Total)
SAMPLING FOR MODEL 'rstanString' NOW (CHAIN 3).
Iteration: 1 / 166667 [ 0%] (Warmup)
Iteration: 16666 / 166667 [ 9%] (Sampling)
Iteration: 33332 / 166667 [ 19%] (Sampling)
Iteration: 49998 / 166667 [ 29%] (Sampling)
Iteration: 66664 / 166667 [ 39%] (Sampling)
Iteration: 83330 / 166667 [ 49%] (Sampling)
Iteration: 99996 / 166667 [ 59%] (Sampling)
Iteration: 116662 / 166667 [ 69%] (Sampling)
Iteration: 133328 / 166667 [ 79%] (Sampling)
Iteration: 149994 / 166667 [ 89%] (Sampling)
Iteration: 166660 / 166667 [ 99%] (Sampling)
Iteration: 166667 / 166667 [100%] (Sampling)
Elapsed Time: 0.94 seconds (Warm-up)
60.2 seconds (Sampling)
61.14 seconds (Total)
> print(data1.rstan)
Inference for Stan model: rstanString.
3 chains, each with iter=166667; warmup=2000; thin=10;
post-warmup draws per chain=16467, total post-warmup draws=49401.
mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat
sd_A 23.3 0 3.6 16.2 21.0 23.3 25.6 30.6 49401 1
sd_Sites 9.1 0 1.8 5.8 7.9 9.0 10.2 12.9 46636 1
sd_Quads 8.3 0 1.1 6.1 7.6 8.4 9.1 10.4 43701 1
beta[1] 35.4 0 5.2 25.1 32.2 35.5 38.7 45.7 48321 1
beta[2] 73.2 0 5.1 62.9 70.0 73.2 76.5 83.4 48485 1
beta[3] 77.1 0 5.1 66.9 73.8 77.1 80.3 87.3 49093 1
sigma 8.9 0 0.7 7.6 8.3 8.8 9.3 10.5 46375 1
sigma_Sites 10.1 0 3.0 5.5 8.0 9.6 11.6 17.1 46273 1
sigma_Quads 8.5 0 1.3 5.9 7.6 8.4 9.3 11.2 44720 1
lp__ -633.1 0 9.2 -652.2 -639.1 -632.8 -626.8 -616.1 46796 1
Samples were drawn using NUTS(diag_e) at Mon Mar 3 09:19:28 2014.
For each parameter, n_eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor on split chains (at
convergence, Rhat=1).
| JAGS Matrix | JAGS Hierarchical | Stan cellmeans | Stan Matrix | |
|---|---|---|---|---|
| Iterations | 10000 | 10000 | 13000 | 13000 |
| Thinning | 10 | 10 | 10 | 10 |
| Samples | 2100 | 2100 | 3000 | 3000 |
| Maximum ACF | 0.1546 | 0.1482 | 0.0576 | 0.0426 |
| beta1 | 35.46 [25.92 , 46.62] | 35.66 [24.14 , 45.58] | 35.36 [24.59 , 44.82] | 35.46 [25.06 , 45.48] |
| beta2 | 37.79 [24.66 , 52.59] | 37.74 [22.82 , 51.78] | 73.53 [64.23 , 84.11] | 37.70 [23.82 , 52.90] |
| beta3 | 41.74 [27.90 , 56.62] | 41.48 [26.96 , 55.73] | 77.29 [66.37 , 87.25] | 41.85 [27.11 , 56.19] |
| sigma | 8.84 [7.41 , 10.27] | 8.79 [7.45 , 10.35] | 8.81 [7.49 , 10.40] | 8.82 [7.48 , 10.40] |
| sigma.Sites | 8.40 [5.89 , 10.99] | 8.40 [5.87 , 11.14] | 9.69 [5.35 , 16.52] | 9.58 [5.00 , 15.85] |
| sigma.Quads | 9.62 [5.01 , 15.62] | 9.77 [4.99 , 16.02] | 8.47 [5.78 , 10.98] | 8.46 [5.94 , 11.09] |
| Time (user self) | 5.17 | 4.14 | 16.35 | 68.82 |
| Time (system self) | 0.01 | 0.00 | 0.03 | 0.10 |
| Time (elapsed) | 5.19 | 4.14 | 2583.75 | 99.64 |
| Time (user child) | 0.00 | 0.00 | 28.27 | 30.30 |
| Time (system child) | 0.00 | 0.00 | 0.64 | 0.53 |
