Tutorial 9.8b - Randomized Complete Block ANOVA (Bayesian)
14 Jan 2014
If you are completely ontop of the conceptual issues pertaining to Randomized Complete Block (RCB) ANOVA, and just need to use this tutorial in order to learn about RCB ANOVA in R, you are invited to skip down to the section on RCB ANOVA in R.
Overview
You are strongly advised to review the information on the nested design in tutorial 9.8a. I am not going to duplicate the overview here.
RCB ANOVA in R
Scenario and Data
Imagine we has designed an experiment in which we intend to measure a response ($y$) to one of treatments (three levels; 'a1', 'a2' and 'a3'). Unfortunately, the system that we intend to sample is spatially heterogeneous and thus will add a great deal of noise to the data that will make it difficult to detect a signal (impact of treatment).
Thus in an attempt to constrain this variability you decide to apply a design (RCB) in which each of the treatments within each of 35 blocks dispersed randomly throughout the landscape. As this section is mainly about the generation of artificial data (and not specifically about what to do with the data), understanding the actual details are optional and can be safely skipped. Consequently, I have folded (toggled) this section away.
- the number of treatments = 3
- the number of blocks containing treatments = 35
- the mean of the treatments = 40, 70 and 80 respectively
- the variability (standard deviation) between blocks of the same treatment = 12
- the variability (standard deviation) between treatments withing blocks = 5
> library(plyr) > set.seed(1) > nTreat <- 3 > nBlock <- 35 > sigma <- 5 > sigma.block <- 12 > n <- nBlock * nTreat > Block <- gl(nBlock, k = 1) > A <- gl(nTreat, k = 1) > dt <- expand.grid(A = A, Block = Block) > # Xmat <- model.matrix(~Block + A + Block:A, data=dt) > Xmat <- model.matrix(~-1 + Block + A, data = dt) > block.effects <- rnorm(n = nBlock, mean = 40, sd = sigma.block) > A.effects <- c(30, 40) > all.effects <- c(block.effects, A.effects) > lin.pred <- Xmat %*% all.effects > > # OR > Xmat <- cbind(model.matrix(~-1 + Block, data = dt), model.matrix(~-1 + A, data = dt)) > ## Sum to zero block effects > block.effects <- rnorm(n = nBlock, mean = 0, sd = sigma.block) > A.effects <- c(40, 70, 80) > all.effects <- c(block.effects, A.effects) > lin.pred <- Xmat %*% all.effects > > > > ## the quadrat observations (within sites) are drawn from normal distributions with means according to > ## the site means and standard deviations of 5 > y <- rnorm(n, lin.pred, sigma) > data <- data.frame(y = y, expand.grid(A = A, Block = Block)) > head(data) #print out the first six rows of the data set
y A Block 1 37.40 1 1 2 61.47 2 1 3 78.07 3 1 4 30.60 1 2 5 59.00 2 2 6 76.73 3 2
> library(ggplot2) > ggplot(data, aes(y = y, x = A)) + geom_boxplot() + facet_grid(. ~ Block)
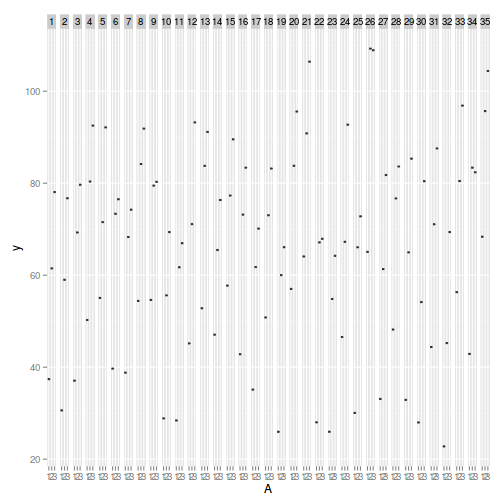
> > library(nlme) > summary(lme(y ~ A, random = ~1 | Block, data))
Linear mixed-effects model fit by REML
Data: data
AIC BIC logLik
722.1 735.2 -356.1
Random effects:
Formula: ~1 | Block
(Intercept) Residual
StdDev: 11.51 4.572
Fixed effects: y ~ A
Value Std.Error DF t-value p-value
(Intercept) 43.03 2.094 68 20.55 0
A2 28.45 1.093 68 26.03 0
A3 40.16 1.093 68 36.74 0
Correlation:
(Intr) A2
A2 -0.261
A3 -0.261 0.500
Standardized Within-Group Residuals:
Min Q1 Med Q3 Max
-1.78748 -0.57868 -0.07108 0.49991 2.33728
Number of Observations: 105
Number of Groups: 35
JAGS
| Full parameterization | Matrix parameterization | Heirarchical parameterization |
|---|---|---|
| $$ \begin{array}{rcl} y_{ijk}&\sim&\mathcal{N}(\mu_{ij},\sigma^2)\\ \mu_{ij} &=& \beta_{j} + \alpha_0 + \alpha_{i} + \\ \beta_{i{j}}&\sim&\mathcal{N}(0,\sigma_{B}^2)\\ \alpha_0, \alpha_i&\sim&\mathcal{N}(0,100000)\\ \sigma^2&\sim&\mathcal{U}(0,100)\\ \sigma_{B}^2&\sim&\mathcal{U}(0,100)\\ \end{array} $$ | $$ \begin{array}{rcl} y_{ijk}&\sim&\mathcal{N}(\mu_{ij},\sigma^2)\\ \mu_{ij} &=& \alpha\mathbf{X} + \beta_{j(i)}\\ \beta_{i{j}}&\sim&\mathcal{N}(0,\sigma_{B}^2)\\ \alpha&\sim&\mathcal{MVN}(0,100000)\\ \sigma^2&\sim&\mathcal{U}(0,100)\\ \sigma_{B}^2&\sim&\mathcal{U}(0,100)\\ \end{array} $$ | $$ \begin{array}{rcl} y_{ij}&\sim &\mathcal{N}(\beta_{j(i)}, \sigma^2)\\ \beta_{j(i)}&\sim&\mathcal{N}(\mu_{i},\sigma_{B}^2)\\ \mu_{i}&=&\alpha\mathbf{X}\\ \alpha_i&\sim&\mathcal{N}(0, 1000000)\\ \sigma^2&\sim&\mathcal{U}(0,100)\\ \sigma_{B}^2&\sim&\mathcal{U}(0,100) \end{array} $$ |
Full effects parameterization
> modelString=" model { #Likelihood for (i in 1:n) { y[i]~dnorm(mu[i],tau) mu[i] <- beta[Block[i]] + alpha0 + alpha[A[i]] } #Priors alpha0 ~ dnorm(0, 1.0E-6) alpha[1] <- 0 for (i in 2:nA) { alpha[i] ~ dnorm(0, 1.0E-6) #prior } for (i in 1:nBlock) { beta[i] ~ dnorm(0, tau.B) #prior } sigma ~ dunif(0, 100) tau <- 1 / (sigma * sigma) sigma.B ~ dunif(0, 100) tau.B <- 1 / (sigma.B * sigma.B) } " > writeLines(modelString,con="../downloads/BUGSscripts/tut9.8jagsF.txt")
> data.list <- with(data, + list(y=y, + Block=as.numeric(Block), + A=as.numeric(A), + n=nrow(data), + nA = length(levels(A)), + nBlock = length(levels(Block)) + ) + ) > > params <- c("alpha0","alpha","sigma","sigma.B") > adaptSteps = 1000 > burnInSteps = 3000 > nChains = 3 > numSavedSteps = 3000 > thinSteps = 10 > nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) > > library(R2jags) > rnorm(1)
[1] -1.524
> jags.f.time <- system.time( + data.r2jags.f <- jags(data=data.list, + inits=NULL, + parameters.to.save=params, + model.file="../downloads/BUGSscripts/tut9.8jagsF.txt", + n.chains=3, + n.iter=nIter, + n.burnin=burnInSteps, + n.thin=thinSteps + ) + )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 472 Initializing model
> print(data.r2jags.f)
Inference for Bugs model at "../downloads/BUGSscripts/tut9.8jagsF.txt", fit using jags,
3 chains, each with 13000 iterations (first 3000 discarded), n.thin = 10
n.sims = 3000 iterations saved
mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff
alpha[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1
alpha[2] 28.439 1.138 26.161 27.691 28.438 29.202 30.641 1.001 3000
alpha[3] 40.165 1.118 37.948 39.426 40.149 40.909 42.353 1.001 3000
alpha0 43.029 2.238 38.681 41.520 42.993 44.534 47.431 1.001 3000
sigma 4.656 0.412 3.927 4.365 4.623 4.919 5.522 1.002 1400
sigma.B 11.902 1.607 9.271 10.745 11.745 12.838 15.600 1.002 1400
deviance 619.505 11.081 600.452 611.733 618.651 626.625 643.985 1.002 1100
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 61.3 and DIC = 680.8
DIC is an estimate of expected predictive error (lower deviance is better).
> data.mcmc.list.f <- as.mcmc(data.r2jags.f)
[1] "alpha[1]" "alpha[2]" "alpha[3]" "alpha0" "deviance" "sigma" "sigma.B"
JAGS Full [1,] "13000" [2,] "10" [3,] "3000" [4,] "0.006" [5,] "42.99 [38.76 , 47.48]" [6,] "28.44 [26.15 , 30.63]" [7,] "40.15 [38.00 , 42.39]" [8,] "4.62 [3.88 , 5.44]" [9,] "11.75 [8.79 , 14.83]" [10,] "4.45" [11,] "0.01" [12,] "4.48" [13,] "0.00" [14,] "0.00"
Matrix parameterization
> modelString=" model { #Likelihood for (i in 1:n) { y[i]~dnorm(mu[i],tau) mu[i] <- beta[Block[i]] + inprod(alpha[],X[i,]) } #Priors alpha ~ dmnorm(a0,A0) for (i in 1:nBlock) { beta[i] ~ dnorm(0, tau.B) #prior } sigma ~ dunif(0, 100) tau <- 1 / (sigma * sigma) sigma.B ~ dunif(0, 100) tau.B <- 1 / (sigma.B * sigma.B) } " > writeLines(modelString,con="../downloads/BUGSscripts/tut9.8jagsM.txt")
> A.Xmat <- model.matrix(~A,data) > data.list <- with(data, + list(y=y, + Block=as.numeric(Block), + X=A.Xmat, + n=nrow(data), + nBlock=length(levels(Block)), + nA = ncol(A.Xmat), + a0=rep(0,3), A0=diag(0,3) + ) + ) > > params <- c("alpha","sigma","sigma.B") > adaptSteps = 1000 > burnInSteps = 3000 > nChains = 3 > numSavedSteps = 3000 > thinSteps = 10 > nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) > > library(R2jags) > rnorm(1)
[1] 1.185
> jags.m.time <- system.time( + data.r2jags.m <- jags(data=data.list, + inits=NULL, + parameters.to.save=params, + model.file="../downloads/BUGSscripts/tut9.8jagsM.txt", + n.chains=3, + n.iter=nIter, + n.burnin=burnInSteps, + n.thin=thinSteps + ) + )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 799 Initializing model
> print(data.r2jags.m)
Inference for Bugs model at "../downloads/BUGSscripts/tut9.8jagsM.txt", fit using jags,
3 chains, each with 13000 iterations (first 3000 discarded), n.thin = 10
n.sims = 3000 iterations saved
mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff
alpha[1] 42.995 2.154 38.750 41.590 42.964 44.425 47.159 1.003 890
alpha[2] 28.441 1.127 26.191 27.675 28.448 29.226 30.564 1.002 1400
alpha[3] 40.186 1.125 37.981 39.440 40.172 40.920 42.431 1.002 1400
sigma 4.671 0.415 3.939 4.387 4.651 4.925 5.553 1.001 3000
sigma.B 11.922 1.581 9.303 10.790 11.762 12.918 15.468 1.001 3000
deviance 619.641 11.264 600.376 611.514 618.868 626.742 643.370 1.001 3000
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 63.5 and DIC = 683.1
DIC is an estimate of expected predictive error (lower deviance is better).
> data.mcmc.list.m <- as.mcmc(data.r2jags.m) > Data.mcmc.list.m <- data.mcmc.list.m
JAGS Full JAGS Matrix [1,] "13000" "13000" [2,] "10" "10" [3,] "3000" "3000" [4,] "0.006" "0.0281" [5,] "42.99 [38.76 , 47.48]" "42.96 [38.66 , 47.05]" [6,] "28.44 [26.15 , 30.63]" "28.45 [26.30 , 30.64]" [7,] "40.15 [38.00 , 42.39]" "40.17 [38.05 , 42.48]" [8,] "4.62 [3.88 , 5.44]" "4.65 [3.90 , 5.49]" [9,] "11.75 [8.79 , 14.83]" "11.76 [9.07 , 15.08]" [10,] "4.45" "4.64" [11,] "0.01" "0.01" [12,] "4.48" "4.66" [13,] "0.00" "0.00" [14,] "0.00" "0.00"
> modelString=" model { #Likelihood (esimating site means (gamma.site) for (i in 1:n) { y[i]~dnorm(mu[i],tau) mu[i] <- beta[Block[i]] + inprod(alpha[],X[i,]) y.err[i] <- y[i] - mu[i] } #Priors alpha ~ dmnorm(a0,A0) for (i in 1:nBlock) { beta[i] ~ dnorm(0, tau.B) #prior } sigma ~ dunif(0, 100) tau <- 1 / (sigma * sigma) sigma.B ~ dunif(0, 100) tau.B <- 1 / (sigma.B * sigma.B) sd.y <- sd(y.err) sd.Block <- sd(beta) sd.A <- sd(alpha) } " > writeLines(modelString,con="../downloads/BUGSscripts/tut9.8bjagsSD.txt")
> A.Xmat <- model.matrix(~A,data) > data.list <- with(data, + list(y=y, + Block=as.numeric(Block), + X=A.Xmat, + n=nrow(data), + nBlock=length(levels(Block)), + nA = ncol(A.Xmat), + a0=rep(0,3), A0=diag(0,3) + ) + ) > > params <- c("alpha","sigma","sd.y",'sd.Block','sd.A','sigma','sigma.B') > adaptSteps = 1000 > burnInSteps = 3000 > nChains = 3 > numSavedSteps = 3000 > thinSteps = 10 > nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) > > library(R2jags) > rnorm(1)
[1] 0.1915
> jags.SD.time <- system.time( + data.r2jagsSD <- jags(data=data.list, + inits=NULL, + parameters.to.save=params, + model.file="../downloads/BUGSscripts/tut9.8bjagsSD.txt", + n.chains=3, + n.iter=nIter, + n.burnin=burnInSteps, + n.thin=thinSteps + ) + )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 909 Initializing model
> print(data.r2jagsSD)
Inference for Bugs model at "../downloads/BUGSscripts/tut9.8bjagsSD.txt", fit using jags,
3 chains, each with 13000 iterations (first 3000 discarded), n.thin = 10
n.sims = 3000 iterations saved
mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff
alpha[1] 43.018 2.210 38.753 41.540 43.010 44.493 47.474 1.001 2500
alpha[2] 28.436 1.106 26.181 27.715 28.445 29.160 30.641 1.003 990
alpha[3] 40.113 1.104 37.937 39.359 40.139 40.830 42.217 1.002 1400
sd.A 6.380 0.876 4.838 5.774 6.309 6.945 8.200 1.002 1700
sd.Block 11.332 0.470 10.404 11.009 11.349 11.660 12.237 1.003 800
sd.y 4.581 0.240 4.182 4.411 4.554 4.722 5.127 1.001 3000
sigma 4.662 0.411 3.937 4.372 4.635 4.917 5.551 1.001 3000
sigma.B 11.925 1.614 9.263 10.772 11.768 12.903 15.543 1.001 3000
deviance 619.303 11.129 599.796 611.638 618.282 626.205 643.523 1.001 3000
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 61.9 and DIC = 681.3
DIC is an estimate of expected predictive error (lower deviance is better).
> data.mcmc.listSD <- as.mcmc(data.r2jagsSD)
RCB (repeated measures) ANOVA in R - continuous within
Scenario and Data
Imagine now that we has designed an experiment to investigate the effects of a continuous predictor ($x$, for example time) on a response ($y$). Again, the system that we intend to sample is spatially heterogeneous and thus will add a great deal of noise to the data that will make it difficult to detect a signal (impact of treatment).
Thus in an attempt to constrain this variability, we again decide to apply a design (RCB) in which each of the levels of X (such as time) treatments within each of 35 blocks dispersed randomly throughout the landscape. As this section is mainly about the generation of artificial data (and not specifically about what to do with the data), understanding the actual details are optional and can be safely skipped. Consequently, I have folded (toggled) this section away.
- the number of times = 10
- the number of blocks containing treatments = 35
- mean slope (rate of change in response over time) = 60
- mean intercept (value of response at time 0 = 200
- the variability (standard deviation) between blocks of the same treatment = 12
- the variability (standard deviation) in slope = 5
> library(plyr) > set.seed(1) > slope <- 30 > intercept <- 200 > nBlock <- 35 > nTime <- 10 > sigma <- 50 > sigma.block <- 30 > n <- nBlock * nTime > Block <- gl(nBlock, k = 1) > Time <- 1:10 > dt <- expand.grid(Time = Time, Block = Block) > Xmat <- model.matrix(~-1 + Block + Time, data = dt) > block.effects <- rnorm(n = nBlock, mean = intercept, sd = sigma.block) > # A.effects <- c(30,40) > all.effects <- c(block.effects, slope) > lin.pred <- Xmat %*% all.effects > > # OR > Xmat <- cbind(model.matrix(~-1 + Block, data = dt), model.matrix(~Time, data = dt)) > ## Sum to zero block effects block.effects <- rnorm(n = nBlock, mean = 0, sd = sigma.block) A.effects > ## <- c(40,70,80) all.effects <- c(block.effects,intercept,slope) lin.pred <- Xmat %*% all.effects > > > > ## the quadrat observations (within sites) are drawn from normal distributions with means according to > ## the site means and standard deviations of 5 > y <- rnorm(n, lin.pred, sigma) > data <- data.frame(y = y, dt) > head(data) #print out the first six rows of the data set
y Time Block 1 190.5 1 1 2 221.5 2 1 3 268.2 3 1 4 356.2 4 1 5 369.4 5 1 6 353.0 6 1
> library(ggplot2) > ggplot(data, aes(y = y, x = Time)) + geom_smooth(method = "lm") + geom_point() + facet_wrap(~Block)
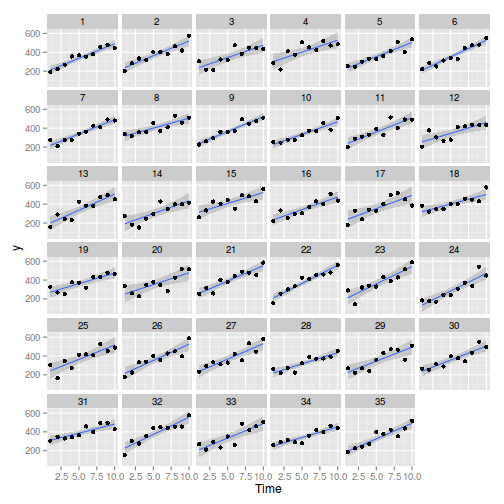
> > library(nlme) > summary(lme(y ~ Time, random = ~1 | Block, data))
Linear mixed-effects model fit by REML
Data: data
AIC BIC logLik
3745 3760 -1868
Random effects:
Formula: ~1 | Block
(Intercept) Residual
StdDev: 21.75 48.23
Fixed effects: y ~ Time
Value Std.Error DF t-value p-value
(Intercept) 209.1 6.674 314 31.33 0
Time 29.1 0.898 314 32.42 0
Correlation:
(Intr)
Time -0.74
Standardized Within-Group Residuals:
Min Q1 Med Q3 Max
-2.71688 -0.67072 -0.01588 0.65160 2.51616
Number of Observations: 350
Number of Groups: 35
> summary(lme(y ~ Time, random = ~1 | Block, data, correlation = corAR1()))
Linear mixed-effects model fit by REML
Data: data
AIC BIC logLik
3744 3764 -1867
Random effects:
Formula: ~1 | Block
(Intercept) Residual
StdDev: 22.81 47.78
Correlation Structure: AR(1)
Formula: ~1 | Block
Parameter estimate(s):
Phi
-0.1034
Fixed effects: y ~ Time
Value Std.Error DF t-value p-value
(Intercept) 208.71 6.390 314 32.66 0
Time 29.15 0.825 314 35.36 0
Correlation:
(Intr)
Time -0.71
Standardized Within-Group Residuals:
Min Q1 Med Q3 Max
-2.73182 -0.66919 -0.02678 0.65341 2.52291
Number of Observations: 350
Number of Groups: 35
> > data$cTime <- scale(data$Time, scale = FALSE) > summary(lme(y ~ cTime, random = ~1 | Block, data))
Linear mixed-effects model fit by REML
Data: data
AIC BIC logLik
3745 3760 -1868
Random effects:
Formula: ~1 | Block
(Intercept) Residual
StdDev: 21.75 48.23
Fixed effects: y ~ cTime
Value Std.Error DF t-value p-value
(Intercept) 369.2 4.491 314 82.20 0
cTime 29.1 0.898 314 32.42 0
Correlation:
(Intr)
cTime 0
Standardized Within-Group Residuals:
Min Q1 Med Q3 Max
-2.71688 -0.67072 -0.01588 0.65160 2.51616
Number of Observations: 350
Number of Groups: 35
> summary(lme(y ~ cTime, random = ~1 | Block, data, correlation = corAR1()))
Linear mixed-effects model fit by REML
Data: data
AIC BIC logLik
3744 3764 -1867
Random effects:
Formula: ~1 | Block
(Intercept) Residual
StdDev: 22.81 47.78
Correlation Structure: AR(1)
Formula: ~1 | Block
Parameter estimate(s):
Phi
-0.1034
Fixed effects: y ~ cTime
Value Std.Error DF t-value p-value
(Intercept) 369.1 4.502 314 81.98 0
cTime 29.2 0.825 314 35.36 0
Correlation:
(Intr)
cTime 0
Standardized Within-Group Residuals:
Min Q1 Med Q3 Max
-2.73182 -0.66919 -0.02678 0.65341 2.52291
Number of Observations: 350
Number of Groups: 35
JAGS
| Full parameterization | Matrix parameterization | Heirarchical parameterization |
|---|---|---|
| $$ \begin{array}{rcl} y_{ijk}&\sim&\mathcal{N}(\mu_{ij},\sigma^2)\\ \mu_{ij} &=& \beta_{j} + \alpha_0 + \alpha_{i} + \\ \beta_{i{j}}&\sim&\mathcal{N}(0,\sigma_{B}^2)\\ \alpha_0, \alpha_i&\sim&\mathcal{N}(0,100000)\\ \sigma^2&\sim&\mathcal{U}(0,100)\\ \sigma_{B}^2&\sim&\mathcal{U}(0,100)\\ \end{array} $$ | $$ \begin{array}{rcl} y_{ijk}&\sim&\mathcal{N}(\mu_{ij},\sigma^2)\\ \mu_{ij} &=& \alpha\mathbf{X} + \beta_{j(i)}\\ \beta_{i{j}}&\sim&\mathcal{N}(0,\sigma_{B}^2)\\ \alpha&\sim&\mathcal{MVN}(0,100000)\\ \sigma^2&\sim&\mathcal{U}(0,100)\\ \sigma_{B}^2&\sim&\mathcal{U}(0,100)\\ \end{array} $$ | $$ \begin{array}{rcl} y_{ij}&\sim &\mathcal{N}(\beta_{j(i)}, \sigma^2)\\ \beta_{j(i)}&\sim&\mathcal{N}(\mu_{i},\sigma_{B}^2)\\ \mu_{i}&=&\alpha\mathbf{X}\\ \alpha_i&\sim&\mathcal{N}(0, 1000000)\\ \sigma^2&\sim&\mathcal{U}(0,100)\\ \sigma_{B}^2&\sim&\mathcal{U}(0,100) \end{array} $$ |
Full effects parameterization
> modelString=" model { #Likelihood for (i in 1:n) { y[i]~dnorm(mu[i],tau) mu[i] <- beta[Block[i]] + alpha0 + alpha1*Time[i] } #Priors alpha0 ~ dnorm(0, 1.0E-6) alpha1 ~ dnorm(0, 1.0E-6) #prior for (i in 1:nBlock) { beta[i] ~ dnorm(0, tau.B) #prior } sigma ~ dunif(0, 100) tau <- 1 / (sigma * sigma) sigma.B ~ dunif(0, 100) tau.B <- 1 / (sigma.B * sigma.B) } " > writeLines(modelString,con="../downloads/BUGSscripts/tut9.8jagsF.T.txt")
> data.list <- with(data, + list(y=y, + Block=as.numeric(Block), + Time=Time, + n=nrow(data), + nBlock = length(levels(Block)) + ) + ) > > params <- c("alpha0","alpha1","sigma","sigma.B") > adaptSteps = 1000 > burnInSteps = 3000 > nChains = 3 > numSavedSteps = 3000 > thinSteps = 10 > nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) > > library(R2jags) > rnorm(1)
[1] 0.9164
> jags.f.T.time <- system.time( + data.r2jags.f.T <- jags(data=data.list, + inits=NULL, + parameters.to.save=params, + model.file="../downloads/BUGSscripts/tut9.8jagsF.T.txt", + n.chains=3, + n.iter=nIter, + n.burnin=burnInSteps, + n.thin=thinSteps + ) + )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 1459 Initializing model
> print(data.r2jags.f.T)
Inference for Bugs model at "../downloads/BUGSscripts/tut9.8jagsF.T.txt", fit using jags,
3 chains, each with 13000 iterations (first 3000 discarded), n.thin = 10
n.sims = 3000 iterations saved
mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff
alpha0 209.30 6.796 196.06 204.81 209.30 213.71 222.66 1.001 3000
alpha1 29.08 0.891 27.33 28.48 29.08 29.66 30.83 1.001 3000
sigma 48.48 1.935 44.94 47.11 48.38 49.70 52.42 1.001 3000
sigma.B 22.61 4.277 14.98 19.61 22.28 25.24 31.93 1.001 2000
deviance 3708.32 9.560 3691.66 3701.51 3707.30 3713.92 3729.71 1.001 3000
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 45.7 and DIC = 3754.0
DIC is an estimate of expected predictive error (lower deviance is better).
> data.mcmc.list.f.T <- as.mcmc(data.r2jags.f.T)
[1] "alpha0" "alpha1" "deviance" "sigma" "sigma.B"
JAGS Full [1,] "13000" [2,] "10" [3,] "3000" [4,] "0.0015" [5,] "209.30 [196.85 , 223.07]" [6,] "29.08 [27.33 , 30.84]" [7,] "48.38 [44.85 , 52.34]" [8,] "22.28 [14.27 , 30.95]" [9,] "12.34" [10,] "0.00" [11,] "12.40" [12,] "0.00" [13,] "0.00"
Matrix parameterization
> modelString=" model { #Likelihood for (i in 1:n) { y[i]~dnorm(mu[i],tau) mu[i] <- beta[Block[i]] + inprod(alpha[],X[i,]) } #Priors alpha ~ dmnorm(a0,A0) for (i in 1:nBlock) { beta[i] ~ dnorm(0, tau.B) #prior } sigma ~ dunif(0, 100) tau <- 1 / (sigma * sigma) sigma.B ~ dunif(0, 100) tau.B <- 1 / (sigma.B * sigma.B) } " > writeLines(modelString,con="../downloads/BUGSscripts/tut9.8jagsM.T.txt")
> Time.Xmat <- model.matrix(~Time,data) > data.list <- with(data, + list(y=y, + Block=as.numeric(Block), + X=Time.Xmat, + n=nrow(data), + nBlock=length(levels(Block)), + a0=rep(0,2), A0=diag(1.0E-06,2) + ) + ) > > params <- c("alpha","sigma","sigma.B") > adaptSteps = 1000 > burnInSteps = 3000 > nChains = 3 > numSavedSteps = 3000 > thinSteps = 10 > nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) > > library(R2jags) > rnorm(1)
[1] 0.3448
> jags.m.T.time <- system.time( + data.r2jags.m.T <- jags(data=data.list, + inits=NULL, + parameters.to.save=params, + model.file="../downloads/BUGSscripts/tut9.8jagsM.T.txt", + n.chains=3, + n.iter=nIter, + n.burnin=burnInSteps, + n.thin=thinSteps + ) + )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 2165 Initializing model
> print(data.r2jags.m.T)
Inference for Bugs model at "../downloads/BUGSscripts/tut9.8jagsM.T.txt", fit using jags,
3 chains, each with 13000 iterations (first 3000 discarded), n.thin = 10
n.sims = 3000 iterations saved
mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff
alpha[1] 209.03 6.724 196.14 204.61 208.96 213.52 222.06 1.001 2800
alpha[2] 29.12 0.875 27.37 28.54 29.12 29.69 30.83 1.001 3000
sigma 48.47 1.945 44.82 47.15 48.40 49.75 52.58 1.002 1500
sigma.B 22.55 4.348 14.92 19.41 22.31 25.27 31.79 1.002 1400
deviance 3708.41 9.553 3692.00 3701.44 3707.58 3714.36 3728.41 1.001 3000
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 45.6 and DIC = 3754.1
DIC is an estimate of expected predictive error (lower deviance is better).
> data.mcmc.list.m.T <- as.mcmc(data.r2jags.m.T) > Data.mcmc.list.m.T <- data.mcmc.list.m.T
JAGS Full JAGS Matrix [1,] "13000" "13000" [2,] "10" "10" [3,] "3000" "3000" [4,] "0.0015" "0.0351" [5,] "209.30 [196.85 , 223.07]" "208.96 [196.15 , 222.08]" [6,] "29.08 [27.33 , 30.84]" "29.13 [27.48 , 30.89]" [7,] "48.38 [44.85 , 52.34]" "48.40 [44.58 , 52.20]" [8,] "22.28 [14.27 , 30.95]" "22.31 [14.87 , 31.70]" [9,] "12.34" "12.31" [10,] "0.00" "0.01" [11,] "12.40" "12.36" [12,] "0.00" "0.00" [13,] "0.00" "0.00"
Hierarchical parameterization
> modelString=" model{ for(DBlock in 1:NBlock) { RBlock[DBlock] ~ dnorm(meanBlock[DBlock], TBlock) meanBlock[DBlock] <- intercept for(Dobservations in SBlock[DBlock]:(SBlock[DBlock+1]-1)){ y[Dobservations] ~ dnorm(meanobservations[Dobservations], Tobservations) meanobservations[Dobservations] <- RBlock[DBlock] + betaobservations * Xobservations[Dobservations] }#observations }#Block # priors intercept ~ dnorm(0, 1.0E-06) betaobservations ~ dnorm(0, 1.0E-06) TBlock <- pow(SDBlock, -2) SDBlock ~ dunif(0, 100) Tobservations <- pow(SDobservations, -2) SDobservations ~ dunif(0, 100) } # model " > > writeLines(modelString,con="../downloads/BUGSscripts/tut9.8jagsH.T.txt")
> library(glmmBUGS) > a <- glmmBUGS(y~Time, effects='Block', family='gaussian', data=data) > data.list=a$ragged > > params <- c("intercept","betaobservations","SDobservations","SDBlock") > adaptSteps = 1000 > burnInSteps = 3000 > nChains = 3 > numSavedSteps = 3000 > thinSteps = 10 > nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) > > library(R2jags) > rnorm(1)
[1] 1.417
> jags.h.T.time <- system.time( + data.r2jags.h.T <- jags(data=data.list, + inits=NULL, + parameters.to.save=params, + model.file="../downloads/BUGSscripts/tut9.8jagsH.T.txt", + n.chains=3, + n.iter=nIter, + n.burnin=burnInSteps, + n.thin=thinSteps + ) + )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 1143 Initializing model
> print(data.r2jags.h.T)
Inference for Bugs model at "../downloads/BUGSscripts/tut9.8jagsH.T.txt", fit using jags,
3 chains, each with 13000 iterations (first 3000 discarded), n.thin = 10
n.sims = 3000 iterations saved
mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff
SDBlock 22.55 4.365 15.01 19.52 22.21 25.15 32.06 1.001 3000
SDobservations 48.41 1.934 44.76 47.03 48.39 49.69 52.45 1.001 3000
betaobservations 29.11 0.911 27.35 28.48 29.10 29.73 30.89 1.001 2700
intercept 209.01 6.832 196.01 204.48 208.91 213.60 222.09 1.001 3000
deviance 3708.22 9.308 3692.28 3701.77 3707.46 3714.10 3728.59 1.001 3000
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 43.3 and DIC = 3751.5
DIC is an estimate of expected predictive error (lower deviance is better).
> data.mcmc.list.h.T <- as.mcmc(data.r2jags.h.T) > Data.mcmc.list.h.T <- data.mcmc.list.h.T
[1] "SDBlock" "SDobservations" "betaobservations" "deviance" "intercept"
JAGS Full JAGS Matrix JAGS Matrix [1,] "13000" "13000" "13000" [2,] "10" "10" "10" [3,] "3000" "3000" "3000" [4,] "0.0015" "0.0351" "0.0235" [5,] "209.30 [196.85 , 223.07]" "208.96 [196.15 , 222.08]" "208.91 [196.00 , 222.08]" [6,] "29.08 [27.33 , 30.84]" "29.13 [27.48 , 30.89]" "29.10 [27.40 , 30.92]" [7,] "48.38 [44.85 , 52.34]" "48.40 [44.58 , 52.20]" "48.39 [44.92 , 52.55]" [8,] "22.28 [14.27 , 30.95]" "22.31 [14.87 , 31.70]" "22.21 [14.76 , 31.64]" [9,] "12.34" "12.31" "13.26" [10,] "0.00" "0.01" "0.00" [11,] "12.40" "12.36" "13.30" [12,] "0.00" "0.00" "0.00" [13,] "0.00" "0.00" "0.00"
RCB (repeated measures) ANOVA in R - continuous within
Scenario and Data
Following on from the previous design (in which a continuous response, $y$ was measured at 10 Time intervals in each of 35 Blocks), we are now going to introduce a categorical covariate measured within each block.
That is, we are measuring the temporal response under two different treatments ($A$) As this section is mainly about the generation of artificial data (and not specifically about what to do with the data), understanding the actual details are optional and can be safely skipped. Consequently, I have folded (toggled) this section away.
- the number of treatments = 2
- the number of times = 10
- the number of blocks containing treatments = 35
- mean slope (rate of change in response over time) = 60
- mean intercept (value of response at time 0 = 200
- the variability (standard deviation) between treatments within blocks = 20
- the variability (standard deviation) between blocks of the same treatment = 12
- the variability (standard deviation) in slope = 5
> library(plyr) > set.seed(1) > slope <- c(30, 10) > treatments <- c(200, 170) > nBlock <- 35 > nTreat <- 2 > nTime <- 10 > sigma <- 50 > sigma.block <- 30 > n <- nBlock * nTime * nTreat > Block <- gl(nBlock, k = 1) > Treatment <- gl(nTreat, k = 1) > Time <- 1:10 > dt <- expand.grid(Time = Time, Treatment = Treatment, Block = Block) > Xmat <- model.matrix(~-1 + Block + Treatment * Time, data = dt) > block.effects <- rnorm(n = nBlock, mean = treatments[1], sd = sigma.block) > all.effects <- c(block.effects, diff(treatments), slope[1], diff(slope)) > lin.pred <- Xmat %*% all.effects > > # OR > Xmat <- cbind(model.matrix(~-1 + Block, data = dt), model.matrix(~Time, data = dt)) > ## Sum to zero block effects block.effects <- rnorm(n = nBlock, mean = 0, sd = sigma.block) A.effects > ## <- c(40,70,80) all.effects <- c(block.effects,intercept,slope) lin.pred <- Xmat %*% all.effects > > > > ## the quadrat observations (within sites) are drawn from normal distributions with means according to > ## the site means and standard deviations of 5 > y <- rnorm(n, lin.pred, sigma) > data <- data.frame(y = y, dt) > head(data) #print out the first six rows of the data set
y Time Treatment Block 1 190.5 1 1 1 2 221.5 2 1 1 3 268.2 3 1 1 4 356.2 4 1 1 5 369.4 5 1 1 6 353.0 6 1 1
> library(ggplot2) > ggplot(data, aes(y = y, x = Time, group = Treatment)) + geom_smooth(method = "lm") + geom_point() + facet_wrap(~Block)
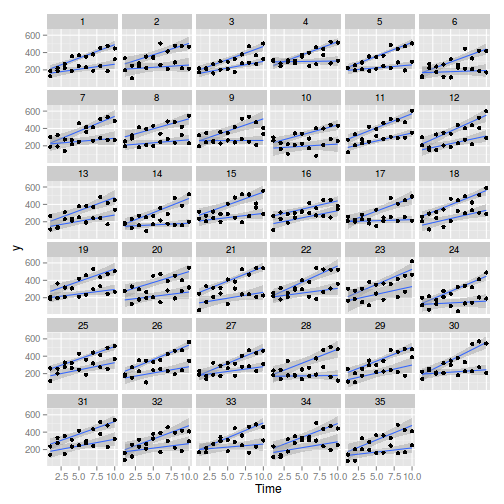
> > library(nlme) > summary(lme(y ~ Time * Treatment, random = ~1 | Block, data))
Linear mixed-effects model fit by REML
Data: data
AIC BIC logLik
7566 7593 -3777
Random effects:
Formula: ~1 | Block
(Intercept) Residual
StdDev: 27.71 51.46
Fixed effects: y ~ Time * Treatment
Value Std.Error DF t-value p-value
(Intercept) 195.37 7.567 662 25.82 0.0000
Time 31.12 0.958 662 32.49 0.0000
Treatment2 -25.33 8.404 662 -3.01 0.0027
Time:Treatment2 -20.98 1.354 662 -15.49 0.0000
Correlation:
(Intr) Time Trtmn2
Time -0.696
Treatment2 -0.555 0.627
Time:Treatment2 0.492 -0.707 -0.886
Standardized Within-Group Residuals:
Min Q1 Med Q3 Max
-2.56257 -0.67946 -0.03499 0.67824 3.51660
Number of Observations: 700
Number of Groups: 35
> summary(lme(y ~ Time * Treatment, random = ~1 | Block, data, correlation = corAR1()))
Linear mixed-effects model fit by REML
Data: data
AIC BIC logLik
7567 7599 -3777
Random effects:
Formula: ~1 | Block
(Intercept) Residual
StdDev: 27.83 51.39
Correlation Structure: AR(1)
Formula: ~1 | Block
Parameter estimate(s):
Phi
-0.02974
Fixed effects: y ~ Time * Treatment
Value Std.Error DF t-value p-value
(Intercept) 195.30 7.470 662 26.14 0.0000
Time 31.14 0.936 662 33.26 0.0000
Treatment2 -25.33 8.186 662 -3.09 0.0021
Time:Treatment2 -21.01 1.319 662 -15.92 0.0000
Correlation:
(Intr) Time Trtmn2
Time -0.689
Treatment2 -0.548 0.623
Time:Treatment2 0.487 -0.705 -0.887
Standardized Within-Group Residuals:
Min Q1 Med Q3 Max
-2.56518 -0.68320 -0.03363 0.68449 3.52429
Number of Observations: 700
Number of Groups: 35
> > data$cTime <- scale(data$Time, scale = FALSE) > summary(lme(y ~ cTime, random = ~1 | Block, data))
Linear mixed-effects model fit by REML
Data: data
AIC BIC logLik
8369 8387 -4181
Random effects:
Formula: ~1 | Block
(Intercept) Residual
StdDev: 21.44 93.9
Fixed effects: y ~ cTime
Value Std.Error DF t-value p-value
(Intercept) 296.17 5.072 664 58.39 0
cTime 20.63 1.236 664 16.70 0
Correlation:
(Intr)
cTime 0
Standardized Within-Group Residuals:
Min Q1 Med Q3 Max
-2.63918 -0.78677 0.01927 0.77208 2.39174
Number of Observations: 700
Number of Groups: 35
> summary(lme(y ~ cTime, random = ~1 | Block, data, correlation = corAR1()))
Linear mixed-effects model fit by REML
Data: data
AIC BIC logLik
7965 7988 -3978
Random effects:
Formula: ~1 | Block
(Intercept) Residual
StdDev: 0.01364 101.8
Correlation Structure: AR(1)
Formula: ~1 | Block
Parameter estimate(s):
Phi
0.7234
Fixed effects: y ~ cTime
Value Std.Error DF t-value p-value
(Intercept) 287.73 8.549 664 33.66 0
cTime 28.34 1.236 664 22.92 0
Correlation:
(Intr)
cTime 0
Standardized Within-Group Residuals:
Min Q1 Med Q3 Max
-2.8993 -0.6157 0.2259 0.8231 2.0782
Number of Observations: 700
Number of Groups: 35
JAGS
| Full parameterization | Matrix parameterization | Heirarchical parameterization |
|---|---|---|
| $$ \begin{array}{rcl} y_{ijk}&\sim&\mathcal{N}(\mu_{ij},\sigma^2)\\ \mu_{ij} &=& \beta_{j} + \alpha_0 + \alpha_{i} + \\ \beta_{i{j}}&\sim&\mathcal{N}(0,\sigma_{B}^2)\\ \alpha_0, \alpha_i&\sim&\mathcal{N}(0,100000)\\ \sigma^2&\sim&\mathcal{U}(0,100)\\ \sigma_{B}^2&\sim&\mathcal{U}(0,100)\\ \end{array} $$ | $$ \begin{array}{rcl} y_{ijk}&\sim&\mathcal{N}(\mu_{ij},\sigma^2)\\ \mu_{ij} &=& \alpha\mathbf{X} + \beta_{j(i)}\\ \beta_{i{j}}&\sim&\mathcal{N}(0,\sigma_{B}^2)\\ \alpha&\sim&\mathcal{MVN}(0,100000)\\ \sigma^2&\sim&\mathcal{U}(0,100)\\ \sigma_{B}^2&\sim&\mathcal{U}(0,100)\\ \end{array} $$ | $$ \begin{array}{rcl} y_{ij}&\sim &\mathcal{N}(\beta_{j(i)}, \sigma^2)\\ \beta_{j(i)}&\sim&\mathcal{N}(\mu_{i},\sigma_{B}^2)\\ \mu_{i}&=&\alpha\mathbf{X}\\ \alpha_i&\sim&\mathcal{N}(0, 1000000)\\ \sigma^2&\sim&\mathcal{U}(0,100)\\ \sigma_{B}^2&\sim&\mathcal{U}(0,100) \end{array} $$ |
Matrix parameterization
> modelString=" model { #Likelihood for (i in 1:n) { y[i]~dnorm(mu[i],tau) mu[i] <- beta[Block[i]] + inprod(alpha[],X[i,]) } #Priors alpha ~ dmnorm(a0,A0) for (i in 1:nBlock) { beta[i] ~ dnorm(0, tau.B) #prior } sigma ~ dunif(0, 100) tau <- 1 / (sigma * sigma) sigma.B ~ dunif(0, 100) tau.B <- 1 / (sigma.B * sigma.B) } " > writeLines(modelString,con="../downloads/BUGSscripts/tut9.8jagsM.TT.txt")
> Xmat <- model.matrix(~Time*Treatment,data) > nXmat <- ncol(Xmat) > data.list <- with(data, + list(y=y, + Block=as.numeric(Block), + X=Xmat, + n=nrow(data), + nBlock=length(levels(Block)), + a0=rep(0,nXmat), A0=diag(1.0E-06,nXmat) + ) + ) > > params <- c("alpha","sigma","sigma.B") > adaptSteps = 1000 > burnInSteps = 3000 > nChains = 3 > numSavedSteps = 3000 > thinSteps = 10 > nIter = burnInSteps+ceiling((numSavedSteps * thinSteps)/nChains) > > library(R2jags) > rnorm(1)
[1] 0.9598
> jags.m.TT.time <- system.time( + data.r2jags.m.TT <- jags(data=data.list, + inits=NULL, + parameters.to.save=params, + model.file="../downloads/BUGSscripts/tut9.8jagsM.TT.txt", + n.chains=3, + n.iter=nIter, + n.burnin=burnInSteps, + n.thin=thinSteps + ) + )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 5689 Initializing model
> print(data.r2jags.m.TT)
Inference for Bugs model at "../downloads/BUGSscripts/tut9.8jagsM.TT.txt", fit using jags,
3 chains, each with 13000 iterations (first 3000 discarded), n.thin = 10
n.sims = 3000 iterations saved
mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff
alpha[1] 195.67 7.723 180.82 190.30 195.72 200.94 210.470 1.001 2500
alpha[2] 31.10 0.944 29.28 30.47 31.11 31.75 32.965 1.001 3000
alpha[3] -25.65 8.475 -41.98 -31.31 -25.56 -20.11 -8.774 1.001 2700
alpha[4] -20.93 1.363 -23.54 -21.83 -20.95 -20.01 -18.261 1.001 3000
sigma 51.54 1.393 48.91 50.60 51.50 52.47 54.476 1.001 3000
sigma.B 28.68 4.215 21.37 25.76 28.25 31.18 38.160 1.001 3000
deviance 7504.94 9.307 7489.16 7498.14 7504.19 7510.81 7525.832 1.002 1600
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 43.3 and DIC = 7548.2
DIC is an estimate of expected predictive error (lower deviance is better).
> data.mcmc.list.m.TT <- as.mcmc(data.r2jags.m.TT) > Data.mcmc.list.m.TT <- data.mcmc.list.m.TT
JAGS Matrix [1,] "13000" [2,] "10" [3,] "3000" [4,] "0.0131" [5,] "195.72 [181.47 , 210.88]" [6,] "31.11 [29.18 , 32.85]" [7,] "-25.56 [-42.04 , -9.05]" [8,] "-20.95 [-23.54 , -18.28]" [9,] "51.50 [48.74 , 54.19]" [10,] "28.25 [20.40 , 37.12]" [11,] "27.82" [12,] "0.02" [13,] "27.94" [14,] "0.00" [15,] "0.00"
