Workshop 9.6b - Factorial ANOVA (Bayesian)
23 April 2011
Factorial ANOVA references
- McCarthy (2007) - Chpt 6
- Kery (2010) - Chpt 10
- Gelman & Hill (2007) - Chpt 4
- Logan (2010) - Chpt 12
- Quinn & Keough (2002) - Chpt 9
> library(R2jags) > library(ggplot2) > library(grid) > #define my common ggplot options > murray_opts <- opts(panel.grid.major=theme_blank(), + panel.grid.minor=theme_blank(), + panel.border = theme_blank(), + panel.background = theme_blank(), + axis.title.y=theme_text(size=15, vjust=0,angle=90), + axis.text.y=theme_text(size=12), + axis.title.x=theme_text(size=15, vjust=-1), + axis.text.x=theme_text(size=12), + axis.line = theme_segment(), + legend.position=c(1,0.1),legend.justification=c(1,0), + legend.key=theme_blank(), + plot.margin=unit(c(0.5,0.5,1,2),"lines") + ) > > #create a convenience function for predicting from mcmc objects > predict.mcmc <- function(mcmc, terms=NULL, mmat, trans="identity") { + library(plyr) + library(coda) + if(is.null(terms)) mcmc.coefs <- mcmc + else { + iCol<-match(terms, colnames(mcmc)) + mcmc.coefs <- mcmc[,iCol] + } + t2=")" + if (trans=="identity") { + t1 <- "" + t2="" + }else if (trans=="log") { + t1="exp(" + }else if (trans=="log10") { + t1="10^(" + }else if (trans=="sqrt") { + t1="(" + t2=")^2" + } + eval(parse(text=paste("mcmc.sum <-adply(mcmc.coefs %*% t(mmat),2,function(x) { data.frame(mean=",t1,"mean(x)",t2,", median=",t1,"median(x)",t2,", ",t1,"coda:::HPDinterval(as.mcmc(x), p=0.68)",t2,", ",t1,"coda:::HPDinterval(as.mcmc(x))",t2," )})", sep=""))) + + mcmc.sum + } > > > #create a convenience function for summarizing mcmc objects > summary.mcmc <- function(mcmc, terms=NULL, trans="identity") { + library(plyr) + library(coda) + if(is.null(terms)) mcmc.coefs <- mcmc + else { + iCol<-grep(terms, colnames(mcmc)) + mcmc.coefs <- mcmc[,iCol] + } + t2=")" + if (trans=="identity") { + t1 <- "" + t2="" + }else if (trans=="log") { + t1="exp(" + }else if (trans=="log10") { + t1="10^(" + }else if (trans=="sqrt") { + t1="(" + t2=")^2" + } + eval(parse(text=paste("mcmc.sum <-adply(mcmc.coefs,2,function(x) { data.frame(mean=",t1,"mean(x)",t2,", median=",t1,"median(x)",t2,", ",t1,"coda:::HPDinterval(as.mcmc(x), p=0.68)",t2,", ",t1,"coda:::HPDinterval(as.mcmc(x))",t2," )})", sep=""))) + + mcmc.sum$Perc <-100*mcmc.sum$median/sum(mcmc.sum$median) + mcmc.sum + }
Two-factor ANOVA
A biologist studying starlings wanted to know whether the mean mass of starlings differed according to different roosting situations. She was also interested in whether the mean mass of starlings altered over winter (Northern hemisphere) and whether the patterns amongst roosting situations were consistent throughout winter, therefore starlings were captured at the start (November) and end of winter (January). Ten starlings were captured from each roosting situation in each season, so in total, 80 birds were captured and weighed.
Download Starling data set| Format of starling.csv data files | |||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
 |
||||||||||||||||||||||||||||||||||||||||||||||||||||
Open the starling data file.
> starling <- read.table("../downloads/data/starling.csv", + header = T, sep = ",", strip.white = T) > head(starling)
SITUATION MONTH MASS GROUP 1 S1 November 78 S1Nov 2 S1 November 88 S1Nov 3 S1 November 87 S1Nov 4 S1 November 88 S1Nov 5 S1 November 83 S1Nov 6 S1 November 82 S1Nov
Exploratory data analysis did not reveal any issues with normality or homogeneity of variance.
- Fit the JAGS model including all pairwise comparisons AND (just because we can), compare the bare surfaces to the surfaces with an algal biofilm. Use with either:
- Effects parameterization
massi = β0 + β1j(i)*monthi + β2j(i)*situationi + β3j(i)*monthi*situationi + εi where ε ∼ N(0,σ2)View full code> modelString=" model { #Likelihood for (i in 1:n) { mass[i]~dnorm(mean[i],tau.res) mean[i] <- beta0+beta.month[month[i]]+beta.situation[situation[i]]+ beta.int[month[i],situation[i]] y.err[i] <- mass[i] - mean[i] } #Priors and derivatives beta0 ~ dnorm(0,1.0E-6) beta.month[1] <- 0 beta.int[1,1] <- 0 for (i in 2:nMonth) { beta.month[i] ~ dnorm(0.001, 1.0E-6) #prior beta.int[i,1] <- 0 } beta.situation[1] <- 0 for (i in 2:nSituation){ beta.situation[i] ~ dnorm(0.001, 1.0E-6) #prior cannot be 0, or else will have an ##init of zero and then will be dividing by zero when calculating cohen's D beta.int[1,i] <- 0 } for (i in 2:nMonth) { for (j in 2:nSituation){ beta.int[i,j] ~ dnorm(0, 1.0E-6) } } #Means for (i in 1:nMonth) { for (j in 1:nSituation) { Mean[i,j] <- beta0 + beta.month[i] + beta.situation[j] + beta.int[i,j] } } sigma.res ~ dgamma(0.001, 0.001) #prior on sd residuals tau.res <- 1 / (sigma.res * sigma.res) sd.Month <- sd(beta.month[]) sd.Situation <- sd(beta.situation[]) sd.Int <- sd(beta.int[,]) sd.Resid <- sd(y.err[]) #Pairwise comparisons of situations for (i in 1:nSituation) { for (j in 1:nSituation) { eff[i,j] <- (beta.situation[i] - beta.situation[j]) } } #Effects of month within each situation for (i in 1:nSituation) { eff2[i] <- Mean[1,i] - Mean[2,i] } } " > ## write the model to a text file (I suggest you alter the path to somewhere more relevant > ## to your system!) > writeLines(modelString,con="../downloads/BUGSscripts/ws9.6bQ1.1a.txt") > starling.list <- with(starling, + list(mass=MASS, + month=as.numeric(MONTH), situation=as.numeric(SITUATION), + n=nrow(starling), nMonth=length(levels(MONTH)), + nSituation=length(levels(SITUATION)) + ) + ) > > params <- c("beta0","beta.month","beta.situation","beta.int","sigma.res","sd.Month","sd.Situation","sd.Int","sd.Resid", "Mean","eff","eff2") > adaptSteps = 1000 > burnInSteps = 200 > nChains = 3 > numSavedSteps = 5000 > thinSteps = 10 > nIter = ceiling((numSavedSteps * thinSteps)/nChains) > > library(R2jags) > ## > starling.r2jags <- jags(data=starling.list, + inits=NULL, #or inits=list(inits,inits,inits) # since there are three chains + parameters.to.save=params, + model.file="../downloads/BUGSscripts/ws9.6bQ1.1a.txt", + n.chains=3, + n.iter=nIter, + n.burnin=burnInSteps, + n.thin=thinSteps + )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 367 Initializing model
> print(starling.r2jags)
Inference for Bugs model at "../downloads/BUGSscripts/ws9.6bQ1.1a.txt", fit using jags, 3 chains, each with 16667 iterations (first 200 discarded), n.thin = 10 n.sims = 4941 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff Mean[1,1] 90.770 1.368 88.088 89.864 90.750 91.695 93.470 1.001 4900 Mean[2,1] 83.630 1.342 80.970 82.743 83.623 84.520 86.276 1.001 4900 Mean[1,2] 90.159 1.348 87.482 89.264 90.161 91.045 92.819 1.001 4900 Mean[2,2] 79.401 1.348 76.754 78.510 79.401 80.284 82.053 1.001 4900 Mean[1,3] 88.211 1.348 85.561 87.329 88.206 89.106 90.850 1.001 4200 Mean[2,3] 78.593 1.341 75.931 77.692 78.588 79.492 81.244 1.001 4900 Mean[1,4] 84.206 1.354 81.575 83.272 84.202 85.124 86.843 1.002 2300 Mean[2,4] 75.438 1.353 72.744 74.560 75.443 76.336 78.079 1.001 4900 beta.int[1,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[2,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[1,2] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[2,2] -3.618 2.741 -9.114 -5.424 -3.608 -1.854 1.970 1.001 4900 beta.int[1,3] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[2,3] -2.478 2.745 -7.906 -4.302 -2.465 -0.639 2.888 1.001 4800 beta.int[1,4] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[2,4] -1.628 2.720 -6.953 -3.497 -1.603 0.259 3.625 1.001 4900 beta.month[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.month[2] -7.140 1.943 -10.964 -8.441 -7.137 -5.837 -3.421 1.001 4900 beta.situation[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.situation[2] -0.611 1.917 -4.434 -1.894 -0.582 0.672 3.168 1.001 4900 beta.situation[3] -2.559 1.900 -6.357 -3.805 -2.557 -1.335 1.222 1.001 2800 beta.situation[4] -6.564 1.938 -10.448 -7.838 -6.562 -5.261 -2.807 1.001 4900 beta0 90.770 1.368 88.088 89.864 90.750 91.695 93.470 1.001 4900 eff[1,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 eff[2,1] -0.611 1.917 -4.434 -1.894 -0.582 0.672 3.168 1.001 4900 eff[3,1] -2.559 1.900 -6.357 -3.805 -2.557 -1.335 1.222 1.001 2800 eff[4,1] -6.564 1.938 -10.448 -7.838 -6.562 -5.261 -2.807 1.001 4900 eff[1,2] 0.611 1.917 -3.168 -0.672 0.582 1.894 4.434 1.001 4900 eff[2,2] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 eff[3,2] -1.948 1.906 -5.640 -3.226 -1.960 -0.673 1.807 1.001 3200 eff[4,2] -5.953 1.897 -9.722 -7.190 -5.958 -4.696 -2.243 1.001 3800 eff[1,3] 2.559 1.900 -1.222 1.335 2.557 3.805 6.357 1.001 2800 eff[2,3] 1.948 1.906 -1.807 0.673 1.960 3.226 5.640 1.001 3200 eff[3,3] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 eff[4,3] -4.005 1.896 -7.681 -5.283 -3.979 -2.766 -0.270 1.002 2000 eff[1,4] 6.564 1.938 2.807 5.261 6.562 7.838 10.448 1.001 4900 eff[2,4] 5.953 1.897 2.243 4.696 5.958 7.190 9.722 1.001 3800 eff[3,4] 4.005 1.896 0.270 2.766 3.979 5.283 7.681 1.002 2000 eff[4,4] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 eff2[1] 7.140 1.943 3.421 5.837 7.137 8.441 10.964 1.004 4900 eff2[2] 10.758 1.931 6.988 9.493 10.754 12.027 14.569 1.001 4900 eff2[3] 9.618 1.918 5.815 8.290 9.647 10.909 13.365 1.001 4900 eff2[4] 8.768 1.878 5.189 7.496 8.742 9.981 12.466 1.002 4900 sd.Int 1.803 0.803 0.516 1.206 1.702 2.306 3.631 1.001 4900 sd.Month 3.570 0.972 1.710 2.918 3.568 4.221 5.482 1.004 4900 sd.Resid 4.185 0.108 4.035 4.106 4.166 4.242 4.446 1.002 1700 sd.Situation 2.741 0.657 1.475 2.296 2.735 3.175 4.089 1.001 3300 sigma.res 4.257 0.358 3.649 4.003 4.235 4.475 5.033 1.001 4900 deviance 457.997 4.558 451.319 454.640 457.201 460.513 468.982 1.002 1700 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 10.4 and DIC = 468.4 DIC is an estimate of expected predictive error (lower deviance is better). - Means parameterization
massi = αj(i)*monthi*situationi + εi where ε ∼ N(0,σ2)View full code
> modelString=" model { #Likelihood for (i in 1:n) { mass[i]~dnorm(mean[i],tau.res) mean[i] <- alpha[month[i],situation[i]] y.err[i] <- mass[i] - mean[i] } #Priors and derivatives for (i in 1:nMonth) { for (j in 1:nSituation){ alpha[i,j] ~ dnorm(0, 1.0E-6) } } sigma.res ~ dgamma(0.001, 0.001) #prior on sd residuals tau.res <- 1 / (sigma.res * sigma.res) #Effects beta0 <- alpha[1,1] for (i in 1:nMonth) { beta.month[i] <- alpha[i,1]-beta0 } for (i in 1:nSituation) { beta.situation[i] <- alpha[1,i]-beta0 } for (i in 1:nMonth) { for (j in 1:nSituation) { beta.int[i,j] <- alpha[i,j]-beta.month[i]-beta.situation[j]-beta0 } } sd.Month <- sd(beta.month[]) sd.Situation <- sd(beta.situation[]) sd.Int <- sd(beta.int[,]) sd.Resid <- sd(y.err[]) #Pairwise comparisons of situations for (i in 1:nSituation) { for (j in 1:nSituation) { eff[i,j] <- beta.situation[i] - beta.situation[j] } } #Effects of month within each situation for (i in 1:nSituation) { eff2[i] <- alpha[1,i] - alpha[2,i] } } " > ## write the model to a text file (I suggest you alter the path to somewhere more relevant > ## to your system!) > writeLines(modelString,con="../downloads/BUGSscripts/ws9.6bQ1.1b.txt") > starling.list <- with(starling, + list(mass=MASS, + month=as.numeric(MONTH), situation=as.numeric(SITUATION), + n=nrow(starling), nMonth=length(levels(MONTH)), + nSituation=length(levels(SITUATION)) + ) + ) > > params <- c("beta.month","beta.situation","beta.int","sigma.res","sd.Month","sd.Situation","sd.Int","sd.Resid", "eff","eff2") > adaptSteps = 1000 > burnInSteps = 200 > nChains = 3 > numSavedSteps = 5000 > thinSteps = 10 > nIter = ceiling((numSavedSteps * thinSteps)/nChains) > > library(R2jags) > ## > starling.r2jags2 <- jags(data=starling.list, + inits=NULL, #or inits=list(inits,inits,inits) # since there are three chains + parameters.to.save=params, + model.file="../downloads/BUGSscripts/ws9.6bQ1.1b.txt", + n.chains=3, + n.iter=nIter, + n.burnin=burnInSteps, + n.thin=thinSteps + )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 381 Initializing model
> print(starling.r2jags2)
Inference for Bugs model at "../downloads/BUGSscripts/ws9.6bQ1.1b.txt", fit using jags, 3 chains, each with 16667 iterations (first 200 discarded), n.thin = 10 n.sims = 4941 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff beta.int[1,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[2,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[1,2] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[2,2] -3.566 2.721 -8.826 -5.398 -3.568 -1.710 1.735 1.001 4900 beta.int[1,3] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[2,3] -2.402 2.666 -7.569 -4.189 -2.387 -0.634 2.796 1.002 1100 beta.int[1,4] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[2,4] -1.568 2.725 -6.805 -3.450 -1.564 0.321 3.712 1.002 1500 beta.month[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.month[2] -7.203 1.919 -10.918 -8.490 -7.208 -5.906 -3.403 1.002 2200 beta.situation[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.situation[2] -0.648 1.921 -4.359 -1.946 -0.637 0.616 3.119 1.001 4900 beta.situation[3] -2.604 1.892 -6.204 -3.883 -2.642 -1.322 1.198 1.001 4900 beta.situation[4] -6.639 1.913 -10.306 -7.943 -6.645 -5.333 -2.831 1.002 2300 eff[1,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 eff[2,1] -0.648 1.921 -4.359 -1.946 -0.637 0.616 3.119 1.001 4900 eff[3,1] -2.604 1.892 -6.204 -3.883 -2.642 -1.322 1.198 1.001 4900 eff[4,1] -6.639 1.913 -10.306 -7.943 -6.645 -5.333 -2.831 1.002 2300 eff[1,2] 0.648 1.921 -3.119 -0.616 0.637 1.946 4.359 1.001 4900 eff[2,2] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 eff[3,2] -1.956 1.914 -5.707 -3.223 -1.957 -0.648 1.726 1.001 4900 eff[4,2] -5.991 1.894 -9.677 -7.263 -5.999 -4.747 -2.186 1.002 1800 eff[1,3] 2.604 1.892 -1.198 1.322 2.642 3.883 6.204 1.001 4900 eff[2,3] 1.956 1.914 -1.726 0.648 1.957 3.223 5.707 1.001 4900 eff[3,3] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 eff[4,3] -4.035 1.904 -7.732 -5.325 -4.031 -2.746 -0.327 1.001 3900 eff[1,4] 6.639 1.913 2.831 5.333 6.645 7.943 10.306 1.002 2300 eff[2,4] 5.991 1.894 2.186 4.747 5.999 7.263 9.677 1.002 1800 eff[3,4] 4.035 1.904 0.327 2.746 4.031 5.325 7.732 1.001 3900 eff[4,4] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 eff2[1] 7.203 1.919 3.403 5.906 7.208 8.490 10.918 1.001 3300 eff2[2] 10.769 1.898 7.117 9.512 10.798 12.029 14.455 1.001 4900 eff2[3] 9.605 1.904 5.859 8.338 9.627 10.853 13.318 1.004 1200 eff2[4] 8.772 1.916 5.019 7.474 8.779 10.084 12.475 1.001 4400 sd.Int 1.779 0.787 0.508 1.195 1.698 2.287 3.499 1.001 4900 sd.Month 3.602 0.960 1.701 2.953 3.604 4.245 5.459 1.001 3300 sd.Resid 4.184 0.104 4.037 4.108 4.167 4.237 4.431 1.001 4900 sd.Situation 2.766 0.644 1.503 2.339 2.768 3.199 4.013 1.002 1800 sigma.res 4.253 0.358 3.626 4.001 4.229 4.476 5.011 1.002 1400 deviance 458.035 4.463 451.313 454.807 457.362 460.524 468.601 1.001 4900 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 10.0 and DIC = 468.0 DIC is an estimate of expected predictive error (lower deviance is better). - Before fully exploring the parameters, it is prudent to examine the convergence and mixing diagnostics.
Chose either the effects or means parameterization model (they should yield much the same).
- Trace plots
View code
> plot(as.mcmc(starling.r2jags))
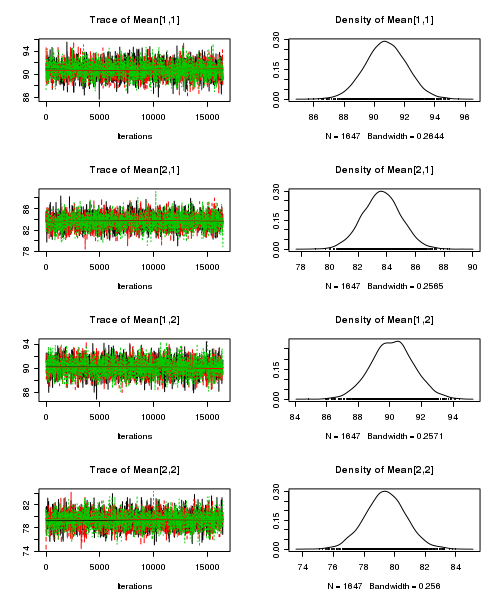
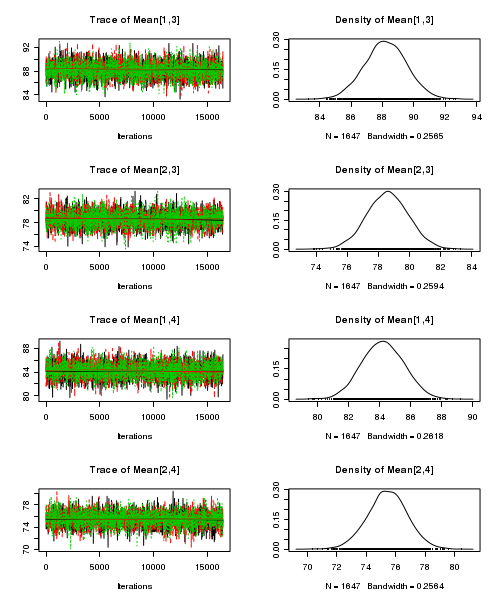
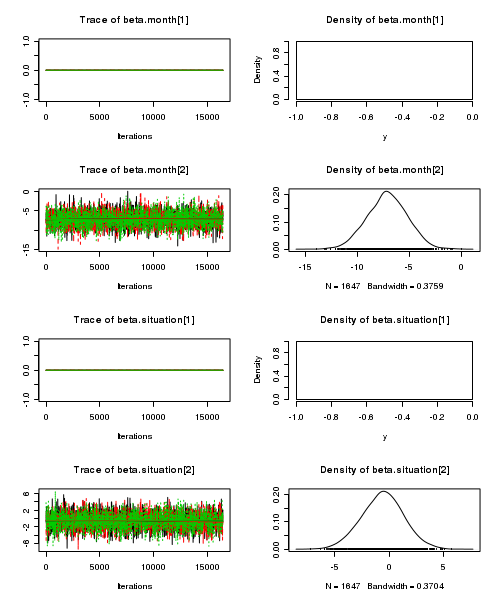
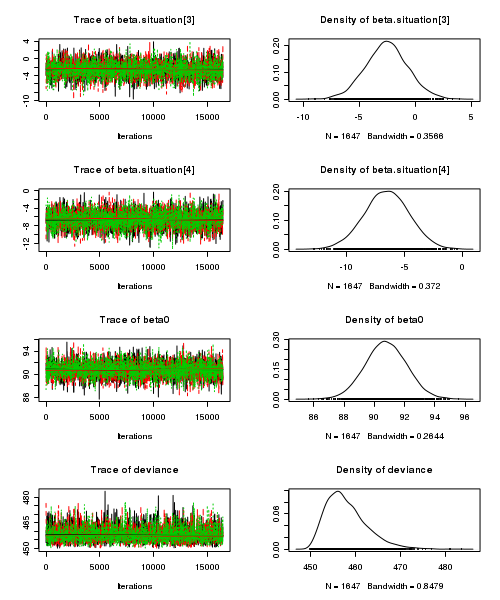
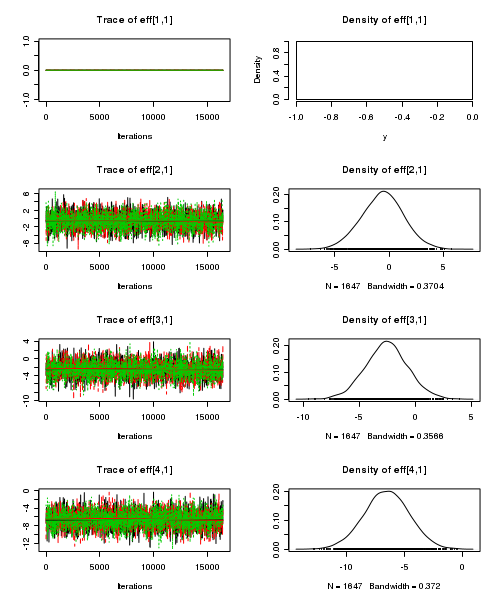
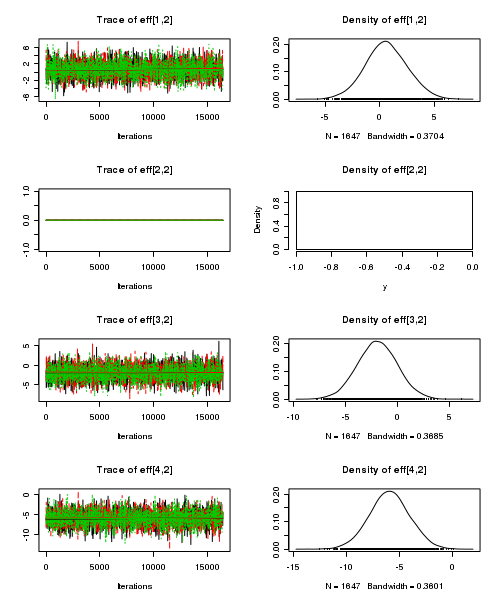

- Raftery diagnostic
View code
> raftery.diag(as.mcmc(starling.r2jags))
[[1]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[2]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[3]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
- Autocorrelation
View code
> autocorr.diag(as.mcmc(starling.r2jags))
Mean[1,1] Mean[2,1] Mean[1,2] Mean[2,2] Mean[1,3] Mean[2,3] Mean[1,4] Mean[2,4] Lag 0 1.000000 1.0000000 1.000000 1.000000 1.0000000 1.000000 1.0000000 1.000000 Lag 10 -0.001908 0.0039394 0.007642 -0.003132 0.0008062 -0.009733 0.0015500 0.004218 Lag 50 -0.010543 0.0007551 -0.023796 0.007433 0.0175925 -0.017918 -0.0002732 -0.018483 Lag 100 -0.017432 0.0037170 0.011733 -0.020159 0.0175249 -0.019843 0.0045985 -0.000722 Lag 500 -0.027325 -0.0155207 0.002354 -0.007411 0.0041462 0.002988 -0.0086342 0.011634 beta.int[1,1] beta.int[2,1] beta.int[1,2] beta.int[2,2] beta.int[1,3] beta.int[2,3] Lag 0 NaN NaN NaN 1.000000 NaN 1.000000 Lag 10 NaN NaN NaN 0.004752 NaN 0.003448 Lag 50 NaN NaN NaN -0.002842 NaN -0.024150 Lag 100 NaN NaN NaN -0.017860 NaN -0.004386 Lag 500 NaN NaN NaN -0.009378 NaN -0.015808 beta.int[1,4] beta.int[2,4] beta.month[1] beta.month[2] beta.situation[1] beta.situation[2] Lag 0 NaN 1.00000 NaN 1.0000000 NaN 1.000000 Lag 10 NaN 0.01550 NaN -0.0045391 NaN 0.007261 Lag 50 NaN 0.01528 NaN 0.0006787 NaN -0.007635 Lag 100 NaN -0.02872 NaN -0.0114574 NaN 0.005118 Lag 500 NaN -0.02084 NaN -0.0224774 NaN -0.003564 beta.situation[3] beta.situation[4] beta0 deviance eff[1,1] eff[2,1] eff[3,1] Lag 0 1.000000 1.00000 1.000000 1.0000000 NaN 1.000000 1.000000 Lag 10 0.001950 0.00691 -0.001908 -0.0098722 NaN 0.007261 0.001950 Lag 50 -0.003427 0.01735 -0.010543 0.0200729 NaN -0.007635 -0.003427 Lag 100 0.004746 -0.01890 -0.017432 0.0006352 NaN 0.005118 0.004746 Lag 500 -0.019668 -0.03489 -0.027325 0.0051727 NaN -0.003564 -0.019668 eff[4,1] eff[1,2] eff[2,2] eff[3,2] eff[4,2] eff[1,3] eff[2,3] eff[3,3] eff[4,3] Lag 0 1.00000 1.000000 NaN 1.000000 1.000000 1.000000 1.000000 NaN 1.00000 Lag 10 0.00691 0.007261 NaN -0.008402 -0.004394 0.001950 -0.008402 NaN 0.01358 Lag 50 0.01735 -0.007635 NaN 0.022240 -0.027287 -0.003427 0.022240 NaN 0.01526 Lag 100 -0.01890 0.005118 NaN 0.008424 0.020021 0.004746 0.008424 NaN 0.02826 Lag 500 -0.03489 -0.003564 NaN -0.019342 0.009797 -0.019668 -0.019342 NaN -0.02784 eff[1,4] eff[2,4] eff[3,4] eff[4,4] eff2[1] eff2[2] eff2[3] eff2[4] sd.Int Lag 0 1.00000 1.000000 1.00000 NaN 1.0000000 1.000000 1.000000 1.000000 1.000000 Lag 10 0.00691 -0.004394 0.01358 NaN -0.0045391 0.007980 0.008606 0.007452 0.007769 Lag 50 0.01735 -0.027287 0.01526 NaN 0.0006787 -0.001802 -0.006377 -0.016473 -0.006266 Lag 100 -0.01890 0.020021 0.02826 NaN -0.0114574 -0.006551 0.008250 -0.003164 -0.025175 Lag 500 -0.03489 0.009797 -0.02784 NaN -0.0224774 -0.015014 0.015986 0.002236 0.005423 sd.Month sd.Resid sd.Situation sigma.res Lag 0 1.0000000 1.000000 1.0000000 1.000000 Lag 10 -0.0045391 -0.001852 -0.0066975 -0.002443 Lag 50 0.0006787 -0.002548 0.0002704 -0.004719 Lag 100 -0.0114574 0.002006 -0.0066467 0.004449 Lag 500 -0.0224774 0.017954 -0.0203869 -0.006040
- Trace plots
- Effects parameterization
-
A summary figure would be nice
View code
> starling.mcmc <- as.mcmc(starling.r2jags$BUGSoutput$sims.matrix) > #get the column indexes for the parameters we are interested in > starling.sum <- summary.mcmc(starling.mcmc, terms=c("Mean")) #or "alpha" > starling.sum <- cbind(starling.sum, expand.grid(Month=c("November","January"), Situation=paste("S",1:4,sep=""))) > > > #starling.sum$name <- factor(rownames(starling.sum), levels=rownames(starling.sum)) > > p1<-ggplot(starling.sum,aes(y=mean, x=Situation, group=Month))+ + geom_errorbar(aes(ymin=lower, ymax=upper),width=0,size=1.5)+ + geom_errorbar(aes(ymin=lower.1, ymax=upper.1),width=0)+ + geom_point(aes(shape=Month, fill=Month),size=3)+ + scale_y_continuous("Starling mass (g)")+ + scale_x_discrete("Roosting situation")+ + scale_shape_manual(name="Month",values=c(21,16), breaks=c("November","January"), labels=c("November","January"))+ + scale_fill_manual(name="Month",values=c("white","black"), breaks=c("November","January"), labels=c("November","January"))+ + murray_opts+opts(legend.position=c(1,0.99),legend.justification=c(1,1)) > > > #Finite-population standard deviations > starling.sd <- summary.mcmc(starling.mcmc, terms=c("sd")) > rownames(starling.sd) <- c("Month:Situation","Month","Residuals", "Situation") > starling.sd$name <- factor(rownames(starling.sd), levels=c("Residuals", "Month:Situation", "Situation","Month")) > > p2<-ggplot(starling.sd,aes(y=name, x=median))+ + geom_vline(xintercept=0,linetype="dashed")+ + geom_hline(xintercept=0)+ + scale_x_continuous("Finite population \nvariance components (sd)")+ + geom_errorbarh(aes(xmin=lower.1, xmax=upper.1), height=0, size=1)+ + geom_errorbarh(aes(xmin=lower, xmax=upper), height=0, size=1.5)+ + geom_point(size=3)+ + geom_text(aes(label=sprintf("(%4.1f%%)",Perc),vjust=-1))+ + murray_opts+ + opts(axis.line=theme_blank(),axis.title.y=theme_blank(), + axis.text.y=theme_text(size=12,hjust=1)) > > grid.newpage() > pushViewport(viewport(layout=grid.layout(1,2,5,5,respect=TRUE))) > pushViewport(viewport(layout.pos.col=1, layout.pos.row=1)) > print(p1,newpage=FALSE) > popViewport(1) > pushViewport(viewport(layout.pos.col=2, layout.pos.row=1)) > print(p2,newpage=FALSE) > popViewport(0)
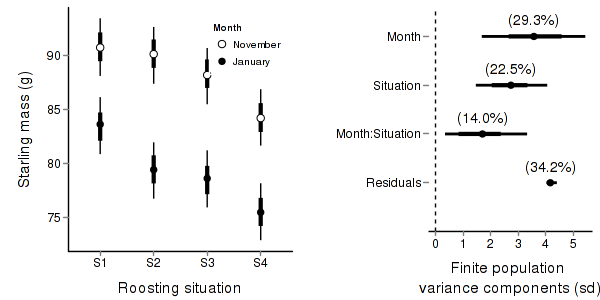
> + + #eff and eff2 > starling.eff <- summary.mcmc(starling.mcmc, terms=c("eff")) > rows <- c(matrix(upper.tri(diag(paste("S",1:4,sep=""))),ncol=1)[,1],rep(TRUE,4)) > nms <- paste("Situation",1:4) > nms <- with(expand.grid(S1=nms, S2=nms),paste(S1,S2,sep=" vs ")) > nms <- c(nms,"Nov - Jan (S1)","Nov - Jan (S2)","Nov - Jan (S3)","Nov - Jan (S4)") > starling.eff$name <- nms > starling.eff <- starling.eff[rows,] > > p3 <- ggplot(starling.eff, aes(y=name, x=mean))+ + geom_vline(xintercept=0,linetype="dashed")+ + geom_hline(xintercept=0)+ + geom_errorbarh(aes(xmin=lower.1, xmax=upper.1), height=0, size=1)+ + geom_errorbarh(aes(xmin=lower, xmax=upper), height=0, size=1.5)+ + geom_point(size=3)+murray_opts+ + scale_x_continuous(name="Effect size")+ + opts(axis.line=theme_blank(),axis.title.y=theme_blank(), + axis.text.y=theme_text(size=12,hjust=1)) > p3
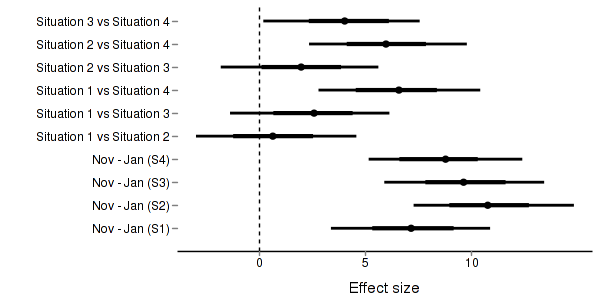
> + + + #cohen's D > eff<-NULL > eff.sd <- NULL > for (i in seq(1,7,b=2)) { + eff <-cbind(eff,starling.mcmc[,i] - starling.mcmc[,i+1]) + eff.sd <- cbind(eff.sd,mean(apply(starling.mcmc[,i:(i+1)],2,sd))) + } > cohen <- sweep(eff,2,eff.sd,"/") > adply(cohen,2,function(x) { + data.frame(mean=mean(x), median=median(x), + HPDinterval(as.mcmc(x), p=0.68), HPDinterval(as.mcmc(x))) + })
Error: unable to find an inherited method for function "HPDinterval", for signature "mcmc"
Unbalanced Two-factor ANOVA
Here is a modified example from Quinn and Keough (2002). Stehman and Meredith (1995) present data from an experiment that was set up to test the hypothesis that healthy spruce seedlings break bud sooner than diseased spruce seedlings. There were 2 factors: pH (3 levels: 3, 5.5, 7) and HEALTH (2 levels: healthy, diseased). The dependent variable was the average (from 5 buds) bud emergence rating (BRATING) on each seedling. The sample size varied for each combination of pH and health, ranging from 7 to 23 seedlings. With two factors, this experiment should be analyzed with a 2 factor (2 x 3) ANOVA.
Download Stehman data set| Format of stehman.csv data files | |||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
 |
||||||||||||||||||||||||||||||||||||||||||||||||||||
Open the stehman data file.
> stehman <- read.table("../downloads/data/stehman.csv", header = T, + sep = ",", strip.white = T) > head(stehman)
PH HEALTH GROUP BRATING 1 3 D D3 0.0 2 3 D D3 0.8 3 3 D D3 0.8 4 3 D D3 0.8 5 3 D D3 0.8 6 3 D D3 0.8
The variable PH contains a list of pH values and is supposed to represent a factorial variable. However, because the contents of this variable are numbers, R initially treats them as numbers, and therefore considers the variable to be numeric rather than categorical. In order to force R/JAGS to treat this variable as a factor (categorical) it must be in categorical form (yet as numbers). Confused? What this means, is that to be treated as a factor, its levels must be indices and therefore the PH variable needs to be converted into a factor before being converted to a numeric. That way the levels 3, 5.5 and 7 will be coded as 1, 2 and 3.
Exploratory data analysis did not reveal any issues with normality or homogeneity of variance.
- Fit the JAGS model including all pairwise comparisons AND (just because we can), compare the bare surfaces to the surfaces with an algal biofilm. Use with either:
- Effects parameterization
bratingi = β0 + β1j(i)*phi + β2j(i)*healthi + β3j(i)*phi*healthi + εi where ε ∼ N(0,σ2)View full code> modelString=" model { #Likelihood for (i in 1:n) { brating[i]~dnorm(mean[i],tau.res) mean[i] <- beta0+beta.ph[ph[i]]+beta.health[health[i]]+ beta.int[ph[i],health[i]] y.err[i] <- brating[i] - mean[i] } #Priors and derivatives beta0 ~ dnorm(0,1.0E-6) beta.ph[1] <- 0 beta.int[1,1] <- 0 for (i in 2:nPh) { beta.ph[i] ~ dnorm(0.001, 1.0E-6) #prior beta.int[i,1] <- 0 } beta.health[1] <- 0 for (i in 2:nHealth){ beta.health[i] ~ dnorm(0.001, 1.0E-6) #prior cannot be 0, or else will have an ##init of zero and then will be dividing by zero when calculating cohen's D beta.int[1,i] <- 0 } for (i in 2:nPh) { for (j in 2:nHealth){ beta.int[i,j] ~ dnorm(0, 1.0E-6) } } #Means for (i in 1:nPh) { for (j in 1:nHealth) { Mean[i,j] <- beta0 + beta.ph[i] + beta.health[j] + beta.int[i,j] } } sigma.res ~ dgamma(0.001, 0.001) #prior on sd residuals tau.res <- 1 / (sigma.res * sigma.res) sd.Ph <- sd(beta.ph[]) sd.Health <- sd(beta.health[]) sd.Int <- sd(beta.int[,]) sd.Resid <- sd(y.err[]) #Pairwise comparisons of PH for (i in 1:nPh) { for (j in 1:nPh) { eff[i,j] <- (beta.ph[i] - beta.ph[j]) eff2[i,j] <- ((Mean[i,1]+Mean[i,2])/2) - ((Mean[j,1]+Mean[j,2])/2) } } # #Effects of ph within each health # for (i in 1:nHealth) { # eff2[i] <- Mean[1,i] - Mean[2,i] # } } " > ## write the model to a text file (I suggest you alter the path to somewhere more relevant > ## to your system!) > writeLines(modelString,con="../downloads/BUGSscripts/ws9.6bQ2.1a.txt") > stehman.list <- with(stehman, + list(brating=BRATING, + ph=as.numeric(as.factor(PH)), health=as.numeric(HEALTH), + n=nrow(stehman), nPh=length(levels(as.factor(PH))), + nHealth=length(levels(HEALTH)) + ) + ) > > params <- c("beta0","beta.ph","beta.health","beta.int","sigma.res","sd.Ph","sd.Health","sd.Int","sd.Resid", "Mean","eff","eff2") > #params <- c("beta0","beta.ph","beta.health","beta.int","sigma.res","Mean", "sd.Ph","sd.Health","sd.Int","sd.Resid", "eff","eff2") > > adaptSteps = 1000 > burnInSteps = 200 > nChains = 3 > numSavedSteps = 5000 > thinSteps = 10 > nIter = ceiling((numSavedSteps * thinSteps)/nChains) > > library(R2jags) > ## > stehman.r2jags <- jags(data=stehman.list, + inits=NULL, #or inits=list(inits,inits,inits) # since there are three chains + parameters.to.save=params, + model.file="../downloads/BUGSscripts/ws9.6bQ2.1a.txt", + n.chains=3, + n.iter=nIter, + n.burnin=burnInSteps, + n.thin=thinSteps + )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 385 Initializing model
> print(stehman.r2jags)
Inference for Bugs model at "../downloads/BUGSscripts/ws9.6bQ2.1a.txt", fit using jags, 3 chains, each with 16667 iterations (first 200 discarded), n.thin = 10 n.sims = 4941 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff Mean[1,1] 1.193 0.107 0.979 1.120 1.194 1.266 1.400 1.001 2700 Mean[2,1] 0.807 0.107 0.592 0.735 0.807 0.879 1.016 1.001 3400 Mean[3,1] 1.125 0.112 0.905 1.050 1.124 1.199 1.348 1.001 4900 Mean[1,2] 1.618 0.156 1.311 1.516 1.616 1.725 1.920 1.001 4900 Mean[2,2] 1.229 0.192 0.849 1.099 1.228 1.359 1.597 1.002 2100 Mean[3,2] 1.337 0.163 1.012 1.228 1.337 1.447 1.656 1.002 2200 beta.health[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.health[2] 0.425 0.189 0.059 0.299 0.425 0.556 0.796 1.002 2400 beta.int[1,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[2,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[3,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[1,2] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[2,2] -0.003 0.292 -0.561 -0.201 -0.001 0.195 0.573 1.001 2600 beta.int[3,2] -0.214 0.273 -0.738 -0.399 -0.211 -0.028 0.333 1.002 1600 beta.ph[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.ph[2] -0.386 0.152 -0.683 -0.488 -0.387 -0.288 -0.081 1.001 4900 beta.ph[3] -0.068 0.156 -0.374 -0.172 -0.069 0.034 0.246 1.001 4700 beta0 1.193 0.107 0.979 1.120 1.194 1.266 1.400 1.001 2700 eff[1,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 eff[2,1] -0.386 0.152 -0.683 -0.488 -0.387 -0.288 -0.081 1.001 4900 eff[3,1] -0.068 0.156 -0.374 -0.172 -0.069 0.034 0.246 1.001 4700 eff[1,2] 0.386 0.152 0.081 0.288 0.387 0.488 0.683 1.001 4900 eff[2,2] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 eff[3,2] 0.318 0.153 0.013 0.215 0.320 0.423 0.621 1.001 4900 eff[1,3] 0.068 0.156 -0.246 -0.034 0.069 0.172 0.374 1.001 4700 eff[2,3] -0.318 0.153 -0.621 -0.423 -0.320 -0.215 -0.013 1.001 4900 eff[3,3] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 eff2[1,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 eff2[2,1] -0.388 0.147 -0.676 -0.486 -0.389 -0.289 -0.099 1.001 4900 eff2[3,1] -0.175 0.135 -0.442 -0.265 -0.175 -0.083 0.091 1.001 4900 eff2[1,2] 0.388 0.147 0.099 0.289 0.389 0.486 0.676 1.001 4900 eff2[2,2] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 eff2[3,2] 0.213 0.149 -0.072 0.114 0.213 0.313 0.504 1.001 4900 eff2[1,3] 0.175 0.135 -0.091 0.083 0.175 0.265 0.442 1.001 4900 eff2[2,3] -0.213 0.149 -0.504 -0.313 -0.213 -0.114 0.072 1.001 4900 eff2[3,3] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 sd.Health 0.214 0.093 0.036 0.150 0.212 0.278 0.398 1.004 1500 sd.Int 0.145 0.074 0.027 0.090 0.137 0.193 0.310 1.001 4900 sd.Ph 0.181 0.059 0.064 0.141 0.181 0.220 0.297 1.001 4900 sd.Resid 0.504 0.009 0.492 0.498 0.502 0.509 0.525 1.001 4900 sigma.res 0.512 0.039 0.442 0.484 0.509 0.537 0.595 1.001 4900 deviance 141.478 3.856 135.933 138.589 140.846 143.614 150.620 1.001 4900 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 7.4 and DIC = 148.9 DIC is an estimate of expected predictive error (lower deviance is better). - Means parameterization
bratingi = αj(i)*phi*healthi + εi where ε ∼ N(0,σ2)View full code
> modelString=" model { #Likelihood for (i in 1:n) { brating[i]~dnorm(mean[i],tau.res) mean[i] <- alpha[ph[i],health[i]] y.err[i] <- brating[i] - mean[i] } #Priors and derivatives for (i in 1:nPh) { for (j in 1:nHealth){ alpha[i,j] ~ dnorm(0, 1.0E-6) } } sigma.res ~ dgamma(0.001, 0.001) #prior on sd residuals tau.res <- 1 / (sigma.res * sigma.res) #Effects beta0 <- alpha[1,1] for (i in 1:nPh) { beta.ph[i] <- alpha[i,1]-beta0 } for (i in 1:nHealth) { beta.health[i] <- alpha[1,i]-beta0 } for (i in 1:nPh) { for (j in 1:nHealth) { beta.int[i,j] <- alpha[i,j]-beta.ph[i]-beta.health[j]-beta0 } } sd.Ph <- sd(beta.ph[]) sd.Health <- sd(beta.health[]) sd.Int <- sd(beta.int[,]) sd.Resid <- sd(y.err[]) #Pairwise comparisons of PH for (i in 1:nPh) { for (j in 1:nPh) { eff[i,j] <- (beta.ph[i] - beta.ph[j]) eff2[i,j] <- ((alpha[i,1]+alpha[i,2])/2) - ((alpha[j,1]+alpha[j,2])/2) } } } " > ## write the model to a text file (I suggest you alter the path to somewhere more relevant > ## to your system!) > writeLines(modelString,con="../downloads/BUGSscripts/ws9.6bQ2.1b.txt") > stehman.list <- with(stehman, + list(brating=BRATING, + ph=as.numeric(as.factor(PH)), health=as.numeric(HEALTH), + n=nrow(stehman), nPh=length(levels(as.factor(PH))), + nHealth=length(levels(HEALTH)) + ) + ) > > params <- c("beta.ph","beta.health","beta.int","sigma.res","sd.Ph","sd.Health","sd.Int","sd.Resid","eff","eff2") > adaptSteps = 1000 > burnInSteps = 200 > nChains = 3 > numSavedSteps = 5000 > thinSteps = 10 > nIter = ceiling((numSavedSteps * thinSteps)/nChains) > > library(R2jags) > ## > stehman.r2jags2 <- jags(data=stehman.list, + inits=NULL, #or inits=list(inits,inits,inits) # since there are three chains + parameters.to.save=params, + model.file="../downloads/BUGSscripts/ws9.6bQ2.1b.txt", + n.chains=3, + n.iter=nIter, + n.burnin=burnInSteps, + n.thin=thinSteps + )
Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 395 Initializing model
> print(stehman.r2jags2)
Inference for Bugs model at "../downloads/BUGSscripts/ws9.6bQ2.1b.txt", fit using jags, 3 chains, each with 16667 iterations (first 200 discarded), n.thin = 10 n.sims = 4941 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff beta.health[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.health[2] 0.430 0.186 0.070 0.305 0.427 0.556 0.791 1.001 4900 beta.int[1,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[2,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[3,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[1,2] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[2,2] -0.008 0.288 -0.581 -0.195 -0.010 0.183 0.556 1.001 4900 beta.int[3,2] -0.215 0.267 -0.739 -0.390 -0.219 -0.034 0.294 1.001 4900 beta.ph[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.ph[2] -0.382 0.151 -0.677 -0.483 -0.382 -0.280 -0.083 1.001 4900 beta.ph[3] -0.067 0.154 -0.368 -0.169 -0.066 0.038 0.233 1.001 4900 eff[1,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 eff[2,1] -0.382 0.151 -0.677 -0.483 -0.382 -0.280 -0.083 1.001 4900 eff[3,1] -0.067 0.154 -0.368 -0.169 -0.066 0.038 0.233 1.001 4900 eff[1,2] 0.382 0.151 0.083 0.280 0.382 0.483 0.677 1.001 4900 eff[2,2] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 eff[3,2] 0.315 0.153 0.017 0.211 0.316 0.417 0.617 1.001 4900 eff[1,3] 0.067 0.154 -0.233 -0.038 0.066 0.169 0.368 1.001 4900 eff[2,3] -0.315 0.153 -0.617 -0.417 -0.316 -0.211 -0.017 1.001 4900 eff[3,3] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 eff2[1,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 eff2[2,1] -0.386 0.146 -0.674 -0.485 -0.384 -0.286 -0.101 1.001 4900 eff2[3,1] -0.174 0.136 -0.442 -0.266 -0.173 -0.083 0.087 1.001 3700 eff2[1,2] 0.386 0.146 0.101 0.286 0.384 0.485 0.674 1.001 4900 eff2[2,2] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 eff2[3,2] 0.212 0.147 -0.072 0.110 0.213 0.309 0.498 1.001 2600 eff2[1,3] 0.174 0.136 -0.087 0.083 0.173 0.266 0.442 1.001 3700 eff2[2,3] -0.212 0.147 -0.498 -0.309 -0.213 -0.110 0.072 1.001 2600 eff2[3,3] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 sd.Health 0.216 0.091 0.039 0.153 0.213 0.278 0.396 1.002 4700 sd.Int 0.143 0.074 0.027 0.089 0.136 0.189 0.307 1.001 3200 sd.Ph 0.179 0.059 0.067 0.138 0.179 0.218 0.296 1.001 4900 sd.Resid 0.504 0.009 0.493 0.498 0.502 0.508 0.526 1.001 4900 sigma.res 0.511 0.038 0.442 0.484 0.509 0.536 0.593 1.001 4900 deviance 141.377 3.820 135.963 138.571 140.734 143.466 150.632 1.001 4900 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 7.3 and DIC = 148.7 DIC is an estimate of expected predictive error (lower deviance is better).
- Effects parameterization
- Before fully exploring the parameters, it is prudent to examine the convergence and mixing diagnostics.
Chose either the effects or means parameterization model (they should yield much the same).
- Trace plots
View code
> plot(as.mcmc(stehman.r2jags))

- Raftery diagnostic
View code
> raftery.diag(as.mcmc(stehman.r2jags))
[[1]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[2]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[3]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
- Autocorrelation
View code
> autocorr.diag(as.mcmc(stehman.r2jags))
Mean[1,1] Mean[2,1] Mean[3,1] Mean[1,2] Mean[2,2] Mean[3,2] beta.health[1] beta.health[2] Lag 0 1.000000 1.00000 1.000000 1.000000 1.0000000 1.000000 NaN 1.000000 Lag 10 0.015825 -0.02483 0.029052 0.001061 -0.0204654 -0.025140 NaN -0.007364 Lag 50 -0.014374 -0.02767 0.003399 0.035938 -0.0004691 -0.003156 NaN -0.002253 Lag 100 0.008493 -0.01021 -0.007150 0.018119 -0.0403369 -0.006234 NaN 0.003267 Lag 500 0.016258 0.02617 -0.019063 0.022172 -0.0003144 0.010743 NaN 0.016014 beta.int[1,1] beta.int[2,1] beta.int[3,1] beta.int[1,2] beta.int[2,2] beta.int[3,2] Lag 0 NaN NaN NaN NaN 1.000000 1.000000 Lag 10 NaN NaN NaN NaN 0.006026 -0.002851 Lag 50 NaN NaN NaN NaN 0.007103 0.000095 Lag 100 NaN NaN NaN NaN -0.006275 -0.012467 Lag 500 NaN NaN NaN NaN 0.007731 0.003171 beta.ph[1] beta.ph[2] beta.ph[3] beta0 deviance eff[1,1] eff[2,1] eff[3,1] eff[1,2] Lag 0 NaN 1.000000 1.000000 1.000000 1.000000 NaN 1.000000 1.000000 1.000000 Lag 10 NaN 0.005331 0.014305 0.015825 -0.006270 NaN 0.005331 0.014305 0.005331 Lag 50 NaN -0.041872 -0.023950 -0.014374 -0.005720 NaN -0.041872 -0.023950 -0.041872 Lag 100 NaN -0.004293 0.001759 0.008493 -0.001357 NaN -0.004293 0.001759 -0.004293 Lag 500 NaN 0.032709 0.003853 0.016258 -0.025478 NaN 0.032709 0.003853 0.032709 eff[2,2] eff[3,2] eff[1,3] eff[2,3] eff[3,3] eff2[1,1] eff2[2,1] eff2[3,1] eff2[1,2] Lag 0 NaN 1.000000 1.000000 1.000000 NaN NaN 1.00000 1.000000 1.00000 Lag 10 NaN -0.001814 0.014305 -0.001814 NaN NaN -0.02276 0.008053 -0.02276 Lag 50 NaN -0.022216 -0.023950 -0.022216 NaN NaN 0.02488 0.008395 0.02488 Lag 100 NaN -0.014981 0.001759 -0.014981 NaN NaN -0.02791 0.009600 -0.02791 Lag 500 NaN 0.012182 0.003853 0.012182 NaN NaN 0.01505 0.031804 0.01505 eff2[2,2] eff2[3,2] eff2[1,3] eff2[2,3] eff2[3,3] sd.Health sd.Int sd.Ph sd.Resid Lag 0 NaN 1.00000 1.000000 1.00000 NaN 1.000000 1.000000 1.000e+00 1.000000 Lag 10 NaN -0.03517 0.008053 -0.03517 NaN -0.007974 -0.007266 4.519e-05 -0.020409 Lag 50 NaN -0.01782 0.008395 -0.01782 NaN 0.001795 0.006918 -2.808e-02 -0.013002 Lag 100 NaN -0.02574 0.009600 -0.02574 NaN 0.001555 -0.025238 -1.349e-02 0.005629 Lag 500 NaN 0.01089 0.031804 0.01089 NaN 0.018169 -0.003348 3.505e-02 -0.042728 sigma.res Lag 0 1.0000000 Lag 10 0.0120803 Lag 50 0.0134666 Lag 100 0.0007495 Lag 500 0.0017084
- Trace plots
-
A summary figure would be nice
View code
> stehman.mcmc <- as.mcmc(stehman.r2jags$BUGSoutput$sims.matrix) > #get the column indexes for the parameters we are interested in > stehman.sum <- summary.mcmc(stehman.mcmc, terms=c("Mean")) #or "alpha" > stehman.sum <- cbind(stehman.sum, expand.grid(Ph=c("3","5.5","7"), Health=c("Diseased","Healthy"))) > > > p1<-ggplot(stehman.sum,aes(y=mean, x=Ph, group=Health))+ + geom_errorbar(aes(ymin=lower, ymax=upper),width=0,size=1.5,position=position_dodge(0.2))+ + geom_errorbar(aes(ymin=lower.1, ymax=upper.1),width=0,position=position_dodge(0.2))+ + geom_point(aes(shape=Health, fill=Health),size=3,position=position_dodge(0.2))+ + scale_y_continuous("Bud emergence rating")+ + scale_x_discrete("Ph")+ + scale_shape_manual(name="Health",values=c(21,16), breaks=c("Diseased","Healthy"), labels=c("Diseased","Healthy"))+ + scale_fill_manual(name="Health",values=c("white","black"), breaks=c("Diseased","Healthy"), labels=c("Diseased","Healthy"))+ + murray_opts+opts(legend.position=c(1,0.99),legend.justification=c(1,1)) > > > #Finite-population standard deviations > stehman.sd <- summary.mcmc(stehman.mcmc, terms=c("sd")) > rownames(stehman.sd) <- c("Health","Ph:Health","Ph", "Residuals") > stehman.sd$name <- factor(c("Health","Ph:Health","Ph", "Residuals"), levels=c("Residuals", "Ph:Health", "Health","Ph")) > > p2<-ggplot(stehman.sd,aes(y=name, x=median))+ + geom_vline(xintercept=0,linetype="dashed")+ + geom_hline(xintercept=0)+ + scale_x_continuous("Finite population \nvariance components (sd)")+ + geom_errorbarh(aes(xmin=lower.1, xmax=upper.1), height=0, size=1)+ + geom_errorbarh(aes(xmin=lower, xmax=upper), height=0, size=1.5)+ + geom_point(size=3)+ + geom_text(aes(label=sprintf("(%4.1f%%)",Perc),vjust=-1))+ + murray_opts+ + opts(axis.line=theme_blank(),axis.title.y=theme_blank(), + axis.text.y=theme_text(size=12,hjust=1)) > > grid.newpage() > pushViewport(viewport(layout=grid.layout(1,2,5,5,respect=TRUE))) > pushViewport(viewport(layout.pos.col=1, layout.pos.row=1)) > print(p1,newpage=FALSE) > popViewport(1) > pushViewport(viewport(layout.pos.col=2, layout.pos.row=1)) > print(p2,newpage=FALSE) > popViewport(0)
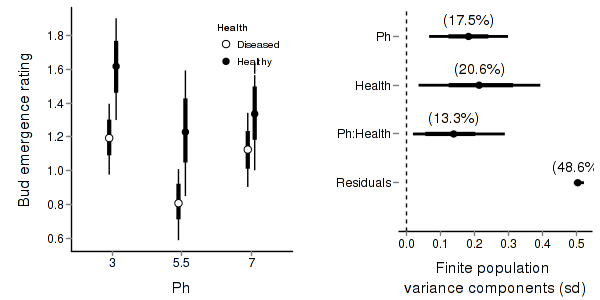
> + + #eff and eff2 > stehman.eff <- summary.mcmc(stehman.mcmc, terms=c("eff\\[")) > rows <- matrix(upper.tri(diag(c("3","5.5","7"))),ncol=1)[,1] > #rows <- c(matrix(upper.tri(diag(c("3","5.5","7"))),ncol=1)[,1], > #matrix(upper.tri(diag(c("3","5.5","7"))),ncol=1)[,1] > #) > nms <- c("3","5.5","7") > nms <- with(expand.grid(S1=nms, S2=nms),paste(S1,S2,sep=" vs ")) > #nms <- c(nms,nms) > stehman.eff$name <- nms > stehman.eff <- stehman.eff[rows,] > > p3 <- ggplot(stehman.eff, aes(y=name, x=mean))+ + geom_vline(xintercept=0,linetype="dashed")+ + geom_hline(xintercept=0)+ + geom_errorbarh(aes(xmin=lower.1, xmax=upper.1), height=0, size=1)+ + geom_errorbarh(aes(xmin=lower, xmax=upper), height=0, size=1.5)+ + geom_point(size=3)+murray_opts+ + scale_x_continuous(name="Effect size")+ + opts(axis.line=theme_blank(),axis.title.y=theme_blank(), + axis.text.y=theme_text(size=12,hjust=1)) > p3
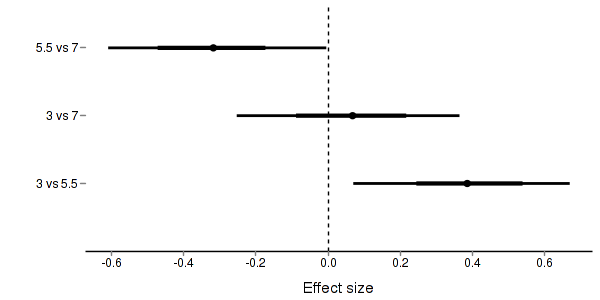
> + + + #cohen's D > eff<-NULL > eff.sd <- NULL > for (i in seq(1,7,b=2)) { + eff <-cbind(eff,stehman.mcmc[,i] - stehman.mcmc[,i+1]) + eff.sd <- cbind(eff.sd,mean(apply(stehman.mcmc[,i:(i+1)],2,sd))) + } > cohen <- sweep(eff,2,eff.sd,"/") > adply(cohen,2,function(x) { + data.frame(mean=mean(x), median=median(x), + HPDinterval(as.mcmc(x), p=0.68), HPDinterval(as.mcmc(x))) + })
Error: unable to find an inherited method for function "HPDinterval", for signature "mcmc"
Two-factor ANOVA with substantial interactions
An ecologist studying a rocky shore at Phillip Island, in southeastern Australia, was interested in how clumps of intertidal mussels are maintained. In particular, he wanted to know how densities of adult mussels affected recruitment of young individuals from the plankton. As with most marine invertebrates, recruitment is highly patchy in time, so he expected to find seasonal variation, and the interaction between season and density - whether effects of adult mussel density vary across seasons - was the aspect of most interest.
The data were collected from four seasons, and with two densities of adult mussels. The experiment consisted of clumps of adult mussels attached to the rocks. These clumps were then brought back to the laboratory, and the number of baby mussels recorded. There were 3-6 replicate clumps for each density and season combination.
Download Quinn data set| Format of quinn.csv data files | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
 |
Open the quinn data file.
> quinn <- read.table("../downloads/data/quinn.csv", header = T, sep = ",", + strip.white = T) > head(quinn)
SEASON DENSITY RECRUITS SQRTRECRUITS GROUP 1 Spring Low 15 3.873 SpringLow 2 Spring Low 10 3.162 SpringLow 3 Spring Low 13 3.606 SpringLow 4 Spring Low 13 3.606 SpringLow 5 Spring Low 5 2.236 SpringLow 6 Spring High 11 3.317 SpringHigh
Exploratory data analysis revealed non-normality and heterogeneous variances (probably due to non-normality). As the response is count data, a square-root transformation is one possible way of addressing these issues.
- Fit the JAGS model including all pairwise comparisons AND (just because we can), compare the bare surfaces to the surfaces with an algal biofilm. Use with either:
- Effects parameterization
recruitsi = β0 + β1j(i)*seasoni + β2j(i)*densityi + β3j(i)*seasoni*densityi + εi where ε ∼ N(0,σ2)View full code> modelString=" data { srecruits <- sqrt(recruits) } model { #Likelihood for (i in 1:n) { srecruits[i]~dnorm(mean[i],tau.res) mean[i] <- beta0+beta.season[season[i]]+beta.density[density[i]]+ beta.int[season[i],density[i]] y.err[i] <- srecruits[i] - mean[i] } #Priors and derivatives beta0 ~ dnorm(0,1.0E-6) beta.season[1] <- 0 beta.int[1,1] <- 0 for (i in 2:nSeason) { beta.season[i] ~ dnorm(0, 1.0E-6) #prior beta.int[i,1] <- 0 } beta.density[1] <- 0 for (i in 2:nDensity){ beta.density[i] ~ dnorm(0, 1.0E-6) #prior cannot be 0, or else will have an beta.int[1,i] <- 0 } for (i in 2:nSeason) { for (j in 2:nDensity){ beta.int[i,j] ~ dnorm(0, 1.0E-6) } } #Means for (i in 1:nSeason) { for (j in 1:nDensity) { Mean[i,j] <- beta0 + beta.season[i] + beta.density[j] + beta.int[i,j] } } sigma.res ~ dgamma(0.001, 0.001) #prior on sd residuals tau.res <- 1 / (sigma.res * sigma.res) sd.Season <- sd(beta.season[]) sd.Density <- sd(beta.density[]) sd.Int <- sd(beta.int[,]) sd.Resid <- sd(y.err[]) #Pairwise comparisons of Density (in each Season) for (i in 1:nSeason) { eff[i] <- Mean[i,1] - Mean[i,2] } } " > ## write the model to a text file (I suggest you alter the path to somewhere more relevant > ## to your system!) > writeLines(modelString,con="../downloads/BUGSscripts/ws9.6bQ3.1a.txt") > quinn.list <- with(quinn, + list(recruits=RECRUITS, + season=as.numeric(SEASON), density=as.numeric(DENSITY), + n=nrow(quinn), nSeason=length(levels(SEASON)), + nDensity=length(levels(DENSITY)) + ) + ) > > #params <- c("beta0","beta.season","beta.density","beta.int","sigma.res","sd.Season","sd.Density","sd.Int","sd.Resid", "Mean","eff","eff2") > params <- c("beta0","beta.season","beta.density","beta.int","sigma.res","Mean", "sd.Season","sd.Density","sd.Int","sd.Resid","eff") > > adaptSteps = 1000 > burnInSteps = 200 > nChains = 3 > numSavedSteps = 5000 > thinSteps = 10 > nIter = ceiling((numSavedSteps * thinSteps)/nChains) > > library(R2jags) > ## > quinn.r2jags <- jags(data=quinn.list, + inits=NULL, #or inits=list(inits,inits,inits) # since there are three chains + parameters.to.save=params, + model.file="../downloads/BUGSscripts/ws9.6bQ3.1a.txt", + n.chains=3, + n.iter=nIter, + n.burnin=burnInSteps, + n.thin=thinSteps + )
Compiling data graph Resolving undeclared variables Allocating nodes Initializing Reading data back into data table Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 249 Initializing model
> print(quinn.r2jags)
Inference for Bugs model at "../downloads/BUGSscripts/ws9.6bQ3.1a.txt", fit using jags, 3 chains, each with 16667 iterations (first 200 discarded), n.thin = 10 n.sims = 4941 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff Mean[1,1] 4.232 0.430 3.409 3.947 4.227 4.520 5.075 1.001 4900 Mean[2,1] 3.023 0.423 2.182 2.752 3.034 3.296 3.857 1.001 4900 Mean[3,1] 6.875 0.430 6.027 6.591 6.877 7.153 7.727 1.001 4900 Mean[4,1] 2.272 0.427 1.437 1.987 2.267 2.559 3.114 1.002 1400 Mean[1,2] 4.264 0.515 3.241 3.927 4.271 4.600 5.275 1.002 1100 Mean[2,2] 3.309 0.465 2.384 2.994 3.308 3.616 4.255 1.001 4900 Mean[3,2] 4.658 0.421 3.820 4.379 4.659 4.935 5.477 1.001 4900 Mean[4,2] 0.925 0.613 -0.318 0.516 0.941 1.338 2.096 1.001 4900 beta.density[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.density[2] 0.032 0.672 -1.260 -0.418 0.037 0.484 1.340 1.002 1700 beta.int[1,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[2,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[3,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[4,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[1,2] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[2,2] 0.254 0.914 -1.562 -0.348 0.250 0.863 2.062 1.002 2300 beta.int[3,2] -2.249 0.911 -4.056 -2.839 -2.253 -1.670 -0.413 1.001 4400 beta.int[4,2] -1.379 0.994 -3.298 -2.044 -1.380 -0.713 0.578 1.002 1200 beta.season[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.season[2] -1.209 0.603 -2.402 -1.620 -1.209 -0.814 -0.040 1.001 4900 beta.season[3] 2.643 0.610 1.437 2.225 2.658 3.047 3.845 1.001 4900 beta.season[4] -1.960 0.604 -3.125 -2.372 -1.958 -1.550 -0.771 1.002 1300 beta0 4.232 0.430 3.409 3.947 4.227 4.520 5.075 1.001 4900 eff[1] -0.032 0.672 -1.340 -0.484 -0.037 0.418 1.260 1.002 1700 eff[2] -0.286 0.620 -1.526 -0.695 -0.276 0.140 0.920 1.001 4900 eff[3] 2.217 0.604 1.036 1.825 2.212 2.613 3.402 1.001 4900 eff[4] 1.347 0.742 -0.134 0.848 1.351 1.845 2.821 1.002 1400 sd.Density 0.269 0.202 0.012 0.111 0.224 0.381 0.758 1.002 4900 sd.Int 0.935 0.270 0.454 0.745 0.922 1.107 1.504 1.001 4900 sd.Resid 1.002 0.053 0.932 0.963 0.992 1.029 1.132 1.001 4900 sd.Season 1.774 0.212 1.360 1.633 1.774 1.911 2.193 1.001 3200 sigma.res 1.033 0.127 0.821 0.942 1.021 1.111 1.317 1.001 4900 deviance 121.313 4.717 114.234 117.829 120.652 124.041 132.017 1.001 4900 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 11.1 and DIC = 132.4 DIC is an estimate of expected predictive error (lower deviance is better). - Means parameterization
srecruitsi = αj(i)*seasoni*densityi + εi where ε ∼ N(0,σ2)View full code
> modelString=" data { srecruits <- sqrt(recruits) } model { #Likelihood for (i in 1:n) { srecruits[i]~dnorm(mean[i],tau.res) mean[i] <- alpha[season[i],density[i]] y.err[i] <- srecruits[i] - mean[i] } #Priors and derivatives for (i in 1:nSeason) { for (j in 1:nDensity){ alpha[i,j] ~ dnorm(0, 1.0E-6) } } sigma.res ~ dgamma(0.001, 0.001) #prior on sd residuals tau.res <- 1 / (sigma.res * sigma.res) #Effects beta0 <- alpha[1,1] for (i in 1:nSeason) { beta.season[i] <- alpha[i,1]-beta0 } for (i in 1:nDensity) { beta.density[i] <- alpha[1,i]-beta0 } for (i in 1:nSeason) { for (j in 1:nDensity) { beta.int[i,j] <- alpha[i,j]-beta.season[i]-beta.density[j]-beta0 } } sd.Season <- sd(beta.season[]) sd.Density <- sd(beta.density[]) sd.Int <- sd(beta.int[,]) sd.Resid <- sd(y.err[]) #Pairwise comparisons of Density (in each Season) for (i in 1:nSeason) { eff[i] <- alpha[i,1] - alpha[i,2] } } " > ## write the model to a text file (I suggest you alter the path to somewhere more relevant > ## to your system!) > writeLines(modelString,con="../downloads/BUGSscripts/ws9.6bQ3.1b.txt") > quinn.list <- with(quinn, + list(recruits=RECRUITS, + season=as.numeric(SEASON), density=as.numeric(DENSITY), + n=nrow(quinn), nSeason=length(levels(SEASON)), + nDensity=length(levels(DENSITY)) + ) + ) > > params <- c("alpha","beta0","beta.season","beta.density","beta.int","sigma.res","sd.Season","sd.Density","sd.Int","sd.Resid","eff") > adaptSteps = 1000 > burnInSteps = 200 > nChains = 3 > numSavedSteps = 5000 > thinSteps = 10 > nIter = ceiling((numSavedSteps * thinSteps)/nChains) > > library(R2jags) > ## > quinn.r2jags2 <- jags(data=quinn.list, + inits=NULL, #or inits=list(inits,inits,inits) # since there are three chains + parameters.to.save=params, + model.file="../downloads/BUGSscripts/ws9.6bQ3.1b.txt", + n.chains=3, + n.iter=nIter, + n.burnin=burnInSteps, + n.thin=thinSteps + )
Compiling data graph Resolving undeclared variables Allocating nodes Initializing Reading data back into data table Compiling model graph Resolving undeclared variables Allocating nodes Graph Size: 263 Initializing model
> print(quinn.r2jags2)
Inference for Bugs model at "../downloads/BUGSscripts/ws9.6bQ3.1b.txt", fit using jags, 3 chains, each with 16667 iterations (first 200 discarded), n.thin = 10 n.sims = 4941 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff alpha[1,1] 4.225 0.426 3.392 3.940 4.228 4.510 5.070 1.001 4900 alpha[2,1] 3.026 0.430 2.200 2.741 3.022 3.317 3.883 1.001 4900 alpha[3,1] 6.859 0.424 6.018 6.581 6.857 7.140 7.703 1.002 2400 alpha[4,1] 2.280 0.428 1.441 1.995 2.283 2.552 3.115 1.001 4700 alpha[1,2] 4.246 0.525 3.212 3.909 4.248 4.600 5.264 1.002 1800 alpha[2,2] 3.285 0.462 2.386 2.974 3.277 3.590 4.206 1.001 4900 alpha[3,2] 4.648 0.422 3.811 4.372 4.649 4.921 5.502 1.001 4900 alpha[4,2] 0.941 0.609 -0.267 0.535 0.944 1.346 2.122 1.001 4900 beta.density[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.density[2] 0.021 0.674 -1.317 -0.412 0.024 0.465 1.337 1.001 3400 beta.int[1,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[2,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[3,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[4,1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[1,2] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.int[2,2] 0.237 0.929 -1.582 -0.378 0.232 0.841 2.062 1.001 4900 beta.int[3,2] -2.232 0.895 -3.975 -2.815 -2.239 -1.666 -0.393 1.001 4900 beta.int[4,2] -1.361 0.995 -3.308 -2.014 -1.361 -0.698 0.564 1.002 2100 beta.season[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1 beta.season[2] -1.198 0.603 -2.367 -1.595 -1.208 -0.799 -0.003 1.001 4900 beta.season[3] 2.635 0.599 1.440 2.261 2.630 3.021 3.816 1.002 3600 beta.season[4] -1.945 0.600 -3.130 -2.353 -1.949 -1.553 -0.787 1.001 4900 beta0 4.225 0.426 3.392 3.940 4.228 4.510 5.070 1.001 4900 eff[1] -0.021 0.674 -1.337 -0.465 -0.024 0.412 1.317 1.001 3400 eff[2] -0.259 0.627 -1.481 -0.679 -0.263 0.156 0.972 1.001 4900 eff[3] 2.211 0.590 1.025 1.820 2.210 2.608 3.370 1.004 1200 eff[4] 1.339 0.735 -0.097 0.862 1.338 1.811 2.785 1.001 4300 sd.Density 0.266 0.207 0.010 0.104 0.220 0.382 0.760 1.001 4900 sd.Int 0.928 0.268 0.451 0.745 0.913 1.090 1.491 1.001 4900 sd.Resid 1.002 0.054 0.929 0.963 0.992 1.029 1.140 1.001 4900 sd.Season 1.765 0.212 1.349 1.628 1.762 1.904 2.193 1.002 1900 sigma.res 1.032 0.129 0.818 0.940 1.019 1.111 1.316 1.001 4900 deviance 121.319 4.838 114.256 117.725 120.497 124.008 133.096 1.001 3300 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 11.7 and DIC = 133.0 DIC is an estimate of expected predictive error (lower deviance is better).
- Effects parameterization
- Before fully exploring the parameters, it is prudent to examine the convergence and mixing diagnostics.
Chose either the effects or means parameterization model (they should yield much the same).
- Trace plots
View code
> plot(as.mcmc(quinn.r2jags))
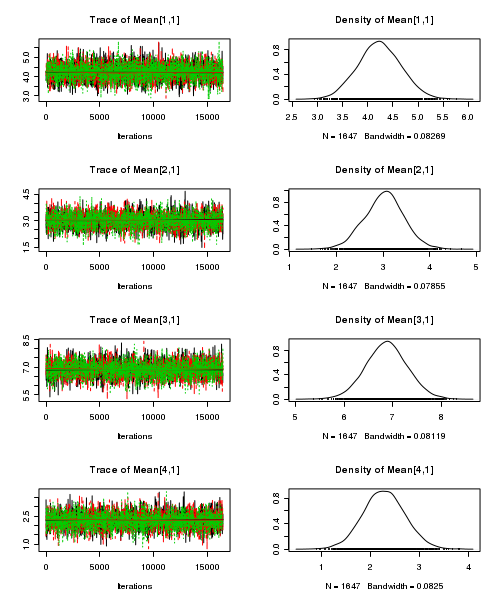
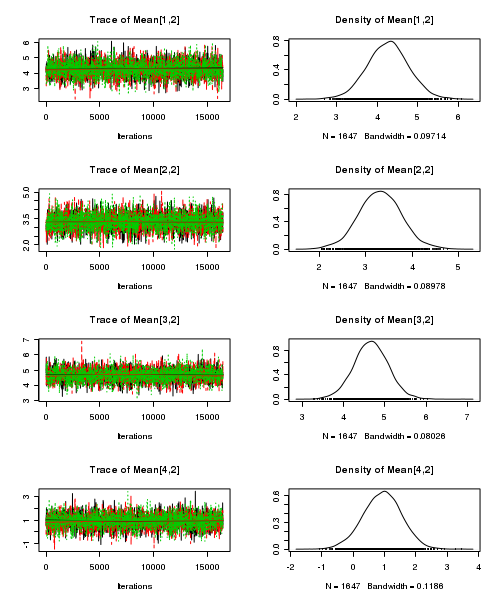

- Raftery diagnostic
View code
> raftery.diag(as.mcmc(quinn.r2jags))
[[1]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[2]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s [[3]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
- Autocorrelation
View code
> autocorr.diag(as.mcmc(quinn.r2jags))
Mean[1,1] Mean[2,1] Mean[3,1] Mean[4,1] Mean[1,2] Mean[2,2] Mean[3,2] Mean[4,2] Lag 0 1.0000000 1.000000 1.000000 1.0000000 1.000000 1.000000 1.000e+00 1.000000 Lag 10 -0.0082297 0.004866 -0.014512 0.0004189 0.019037 0.013411 4.773e-03 -0.008294 Lag 50 0.0008819 -0.003041 -0.005565 0.0035012 0.007405 0.013454 -1.337e-06 0.016589 Lag 100 -0.0060674 -0.009696 -0.006897 -0.0111460 -0.001689 0.014131 -2.677e-03 0.013051 Lag 500 -0.0119590 -0.001043 -0.019341 0.0072319 -0.008894 -0.009759 -2.027e-02 0.002028 beta.density[1] beta.density[2] beta.int[1,1] beta.int[2,1] beta.int[3,1] beta.int[4,1] Lag 0 NaN 1.000000 NaN NaN NaN NaN Lag 10 NaN 0.003179 NaN NaN NaN NaN Lag 50 NaN 0.007616 NaN NaN NaN NaN Lag 100 NaN -0.010429 NaN NaN NaN NaN Lag 500 NaN -0.011344 NaN NaN NaN NaN beta.int[1,2] beta.int[2,2] beta.int[3,2] beta.int[4,2] beta.season[1] beta.season[2] Lag 0 NaN 1.0000000 1.000000 1.000000 NaN 1.000000 Lag 10 NaN 0.0299663 -0.007553 0.005904 NaN -0.011382 Lag 50 NaN 0.0000938 0.003024 -0.005636 NaN -0.006023 Lag 100 NaN 0.0022546 -0.012278 -0.008113 NaN -0.010964 Lag 500 NaN -0.0093048 -0.002738 -0.025463 NaN -0.010620 beta.season[3] beta.season[4] beta0 deviance eff[1] eff[2] eff[3] eff[4] Lag 0 1.00000 1.000000 1.0000000 1.000000 1.000000 1.000000 1.000000 1.000000 Lag 10 -0.02272 -0.002790 -0.0082297 -0.004244 0.003179 0.028275 -0.001557 -0.009585 Lag 50 0.01592 0.004193 0.0008819 0.001471 0.007616 -0.004404 -0.015173 0.006090 Lag 100 -0.01495 -0.017531 -0.0060674 0.008789 -0.010429 0.015171 -0.019009 -0.009054 Lag 500 -0.01460 -0.019725 -0.0119590 0.008385 -0.011344 -0.007830 -0.026246 -0.003047 sd.Density sd.Int sd.Resid sd.Season sigma.res Lag 0 1.000000 1.000000 1.000000 1.0000000 1.000000 Lag 10 -0.016133 -0.017361 -0.013893 0.0028862 0.008276 Lag 50 0.006714 -0.005050 0.017671 -0.0029169 -0.009613 Lag 100 0.019943 0.004759 0.008819 -0.0102643 -0.025626 Lag 500 0.014718 -0.025343 0.013722 0.0006947 -0.016266
- Trace plots
-
A summary figure would be nice
View code
> quinn.mcmc <- as.mcmc(quinn.r2jags$BUGSoutput$sims.matrix) > #get the column indexes for the parameters we are interested in > quinn.sum <- summary.mcmc(quinn.mcmc, terms=c("Mean")) #or "alpha" > quinn.sum <- cbind(quinn.sum, expand.grid(Season=c("Autumn","Spring","Summer","Winter"), Density=c("High","Low"))) > > quinn.sum$Season <- factor(quinn.sum$Season, levels=c("Spring","Summer","Autumn","Winter")) > library(scales) > > p1<-ggplot(quinn.sum,aes(y=mean, x=Season, group=Density))+ + geom_errorbar(aes(ymin=lower, ymax=upper),width=0,size=1.5,position=position_dodge(0.2))+ + geom_errorbar(aes(ymin=lower.1, ymax=upper.1),width=0,position=position_dodge(0.2))+ + geom_point(aes(shape=Density, fill=Density),size=3,position=position_dodge(0.2))+ + scale_y_continuous("Number of mussel recruits", + trans=trans_new("x2",function(x) x^2,"sqrt"), + breaks=sqrt(pretty(c(0,60))), + labels=pretty(c(0,60)))+ + scale_x_discrete("Season")+ + scale_shape_manual(name="Density",values=c(21,16), breaks=c("High","Low"), labels=c("High","Low"))+ + scale_fill_manual(name="Density",values=c("white","black"), breaks=c("High","Low"), labels=c("High","Low"))+ + murray_opts+opts(legend.position=c(1,0.99),legend.justification=c(1,1)) > p1
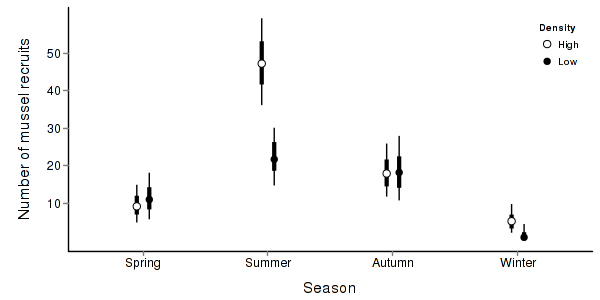
> + + #Finite-population standard deviations > quinn.sd <- summary.mcmc(quinn.mcmc, terms=c("sd")) > rownames(quinn.sd) <- c("Density","Season:Density","Residuals","Season") > quinn.sd$name <- factor(c("Density","Season:Density","Residuals","Season"), levels=c("Residuals", "Season:Density", "Density","Season")) > > p2<-ggplot(quinn.sd,aes(y=name, x=median))+ + geom_vline(xintercept=0,linetype="dashed")+ + geom_hline(xintercept=0)+ + scale_x_continuous("Finite population \nvariance components (sd)")+ + geom_errorbarh(aes(xmin=lower.1, xmax=upper.1), height=0, size=1)+ + geom_errorbarh(aes(xmin=lower, xmax=upper), height=0, size=1.5)+ + geom_point(size=3)+ + geom_text(aes(label=sprintf("(%4.1f%%)",Perc),vjust=-1))+ + murray_opts+ + opts(axis.line=theme_blank(),axis.title.y=theme_blank(), + axis.text.y=theme_text(size=12,hjust=1)) > > grid.newpage() > pushViewport(viewport(layout=grid.layout(1,2,5,5,respect=TRUE))) > pushViewport(viewport(layout.pos.col=1, layout.pos.row=1)) > print(p1,newpage=FALSE) > popViewport(1) > pushViewport(viewport(layout.pos.col=2, layout.pos.row=1)) > print(p2,newpage=FALSE) > popViewport(0)
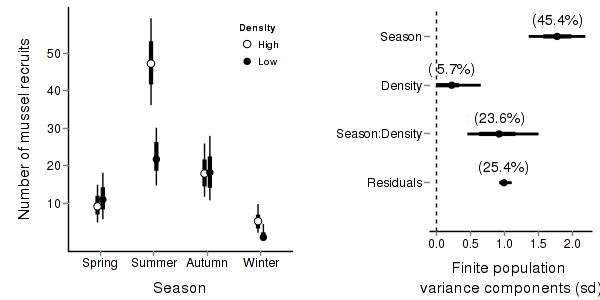
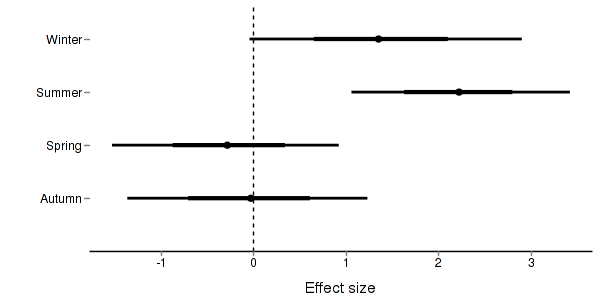
> + + #eff > quinn.eff <- summary.mcmc(quinn.mcmc, terms=c("eff\\[")) > nms <- c("Autumn","Spring","Summer","Winter") > quinn.eff$name <- nms > > p3 <- ggplot(quinn.eff, aes(y=name, x=mean))+ + geom_vline(xintercept=0,linetype="dashed")+ + geom_hline(xintercept=0)+ + geom_errorbarh(aes(xmin=lower.1, xmax=upper.1), height=0, size=1)+ + geom_errorbarh(aes(xmin=lower, xmax=upper), height=0, size=1.5)+ + geom_point(size=3)+murray_opts+ + scale_x_continuous(name="Effect size")+ + opts(axis.line=theme_blank(),axis.title.y=theme_blank(), + axis.text.y=theme_text(size=12,hjust=1)) > p3
> + + + #cohen's D > #eff<-NULL > #eff.sd <- NULL > #for (i in seq(1,7,b=2)) { > # eff <-cbind(eff,quinn.mcmc[,i] - quinn.mcmc[,i+1]) > # eff.sd <- cbind(eff.sd,mean(apply(quinn.mcmc[,i:(i+1)],2,sd))) > #} > #cohen <- sweep(eff,2,eff.sd,"/") > #adply(cohen,2,function(x) { > # data.frame(mean=mean(x), median=median(x), > # HPDinterval(as.mcmc(x), p=0.68), HPDinterval(as.mcmc(x))) > #})
